Characterize batch effects
Almut Lütge
19 April, 2020
pancreas
suppressPackageStartupMessages({
library(CellBench)
library(scater)
library(CellMixS)
library(variancePartition)
library(purrr)
library(jcolors)
library(here)
library(tidyr)
library(dplyr)
library(stringr)
library(ComplexHeatmap)
#library(ggtern)
library(gridExtra)
library(scran)
library(cowplot)
library(CAMERA)
library(ggrepel)
library(readr)
})
options(bitmapType='cairo')Dataset and parameter
sce <- readRDS(params$data)
param <- readRDS(params$param)
celltype <- param[["celltype"]]
batch <- param[["batch"]]
sample <- param[["sample"]]
dataset_name <- param[["dataset_name"]]
dataset_name## [1] "pancreas"n_genes <- nrow(sce)
table(colData(sce)[,celltype])##
## acinar activated_stellate alpha beta
## 690 164 2042 914
## delta ductal endothelial epsilon
## 380 1029 47 13
## gamma macrophage mast quiescent_stellate
## 341 23 14 19
## schwann
## 7table(colData(sce)[,batch])##
## celseq celseq2 smartseq2
## 1004 2285 2394res_de <- readRDS(params$de)
abund <- readRDS(params$abund)
outputfile <- params$out_file
cols <-c(c(jcolors('pal6'),jcolors('pal8'))[c(1,8,14,5,2:4,6,7,9:13,15:20)],jcolors('pal4'))
names(cols) <- c()Visualize data
How are sample, celltypes and batches distributed within normalized, but not batch corrected data?
feature_list <- c(batch, celltype, sample)
feature_list <- feature_list[which(!is.na(feature_list))]
lapply(feature_list, function(feature_name){
visGroup(sce, feature_name, dim_red= "UMAP")
})## [[1]]
##
## [[2]]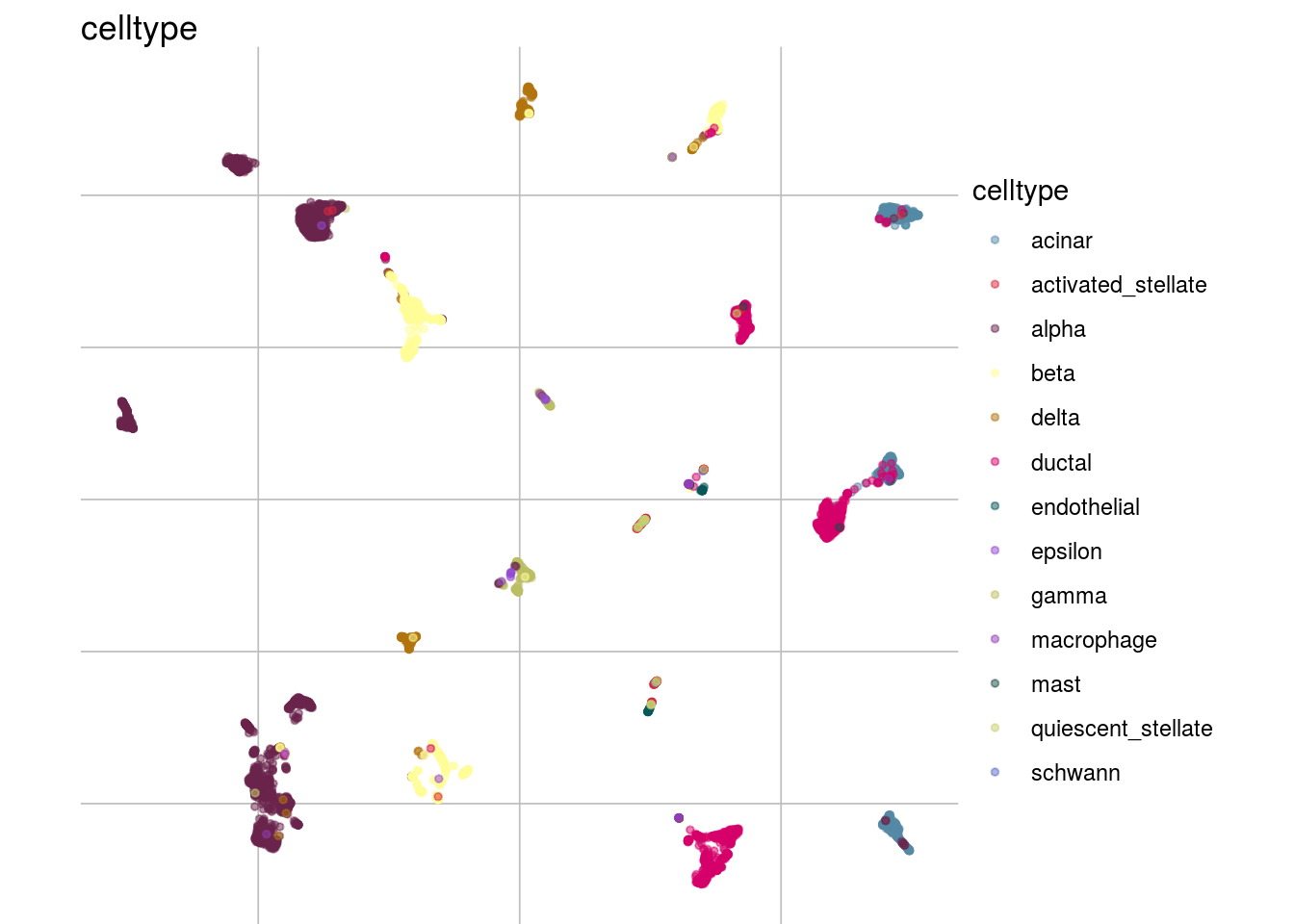
##
## [[3]]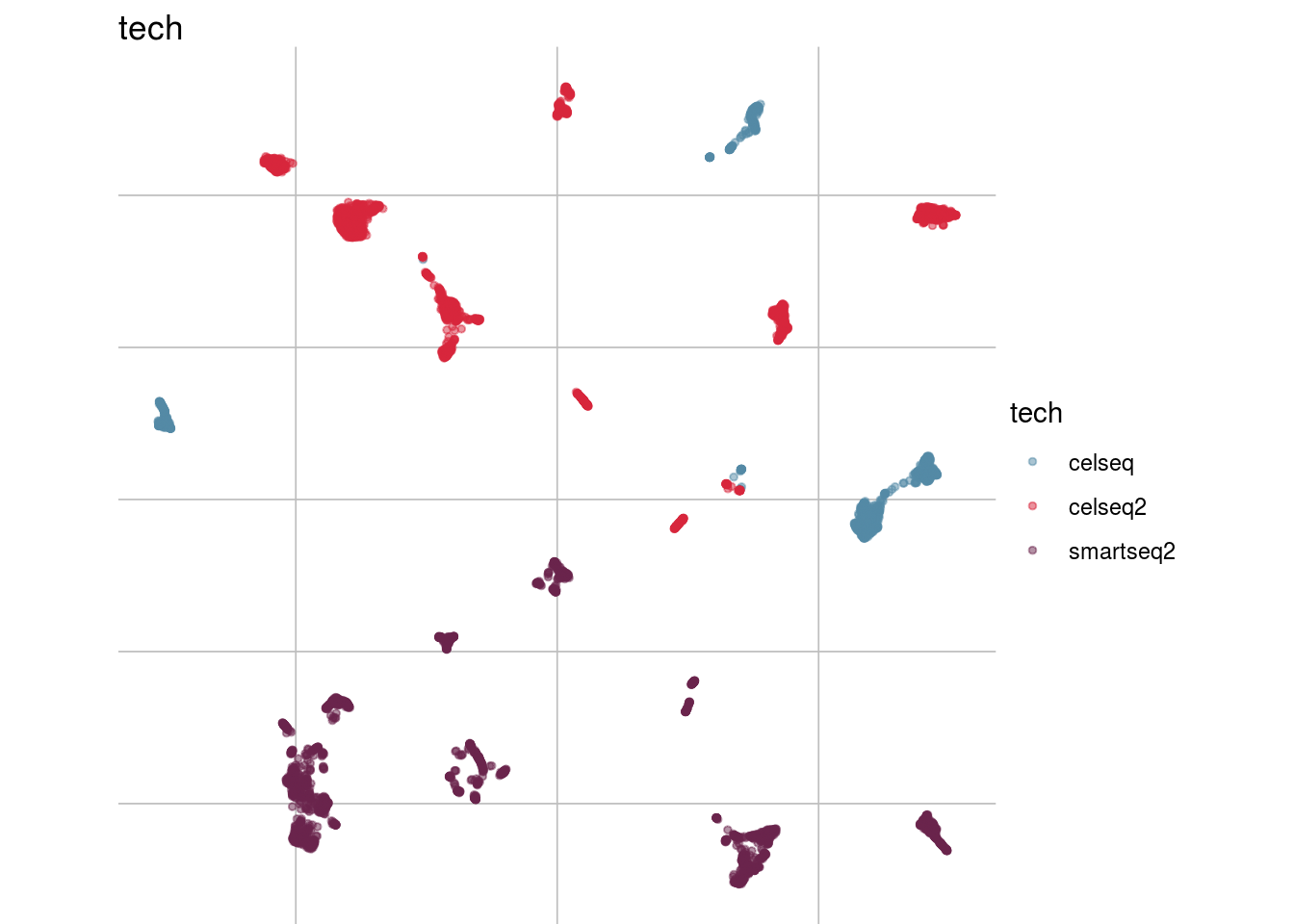
Batch strength/size
To compare or describe the severity of a batch effect there are different meassures. In general they can either give an estimate of the relative strength compared to the signal of interest e.g. the celltype signal or an absolut estimate e.g. the number of batch affected genes.
Variance partitioning
How much of the variance within the datasets can we attributed to the batch effect and how much could be explained by the celltype? Which genes are mostly affected?
vp_vars <- c("vp_batch", "vp_celltype", "vp_residuals")
vp <- as_tibble(rowData(sce)[, vp_vars]) %>% dplyr::mutate(gene= rownames(sce)) %>% dplyr::arrange(-vp_batch)
vp_sub <- vp[1:3] %>% set_rownames(vp$gene)## Warning: Setting row names on a tibble is deprecated.#plot
plotPercentBars( vp_sub[1:10,] )
plotVarPart( vp_sub )
Variance and gene expression
Are general expression and batch effect related? Does the batch effect or the celltype effect preferable manifest within highly, medium or low expressed genes?
#define expression classes by mean expression quantiles
th <- quantile(rowMeans(assays(sce)$logcounts), c(.33, .66))
high_th <- th[2]
mid_th <- th[1]
rowData(sce)$expr_class <- ifelse(rowMeans(assays(sce)$logcounts) > high_th, "high",
ifelse(rowMeans(assays(sce)$logcounts) <= high_th &
rowMeans(assays(sce)$logcounts) > mid_th,
"medium", "low"))
rowData(sce)$mean_expr <- rowMeans(assays(sce)$logcounts)
#plot
plot_dev <- function(var, var_col){
ggplot(as.data.frame(rowData(sce)), aes_string(x = "mean_expr", y = var, colour = var_col)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE)
}
#Ternary plots
# ggtern(data=as.data.frame(rowData(sce)),aes(vp_batch, vp_celltype, vp_residuals)) +
# stat_density_tern(aes(fill=..level.., alpha=..level..),geom='polygon') +
# scale_fill_gradient2(high = "red") +
# guides(color = "none", fill = "none", alpha = "none") +
# geom_point(size= 0.1, alpha = 0.5) +
# Llab("batch") +
# Tlab("celltype") +
# Rlab("other") +
# theme_bw()
#
# t1 <- ggtern(data=as.data.frame(rowData(sce)),aes(vp_batch, vp_celltype, vp_residuals)) +
# geom_point(size = 0.1) +
# geom_density_tern() +
# Llab("batch") +
# Tlab("celltype") +
# Rlab("other") +
# theme_bw()
## Summarize variance partitioning
# How many genes have a variance component affected by batch with > 1%
n_batch_gene <- vp_sub %>% dplyr::filter(vp_batch > 0.01) %>% nrow()/n_genes
n_batch_gene10 <- vp_sub %>% dplyr::filter(vp_batch > 0.1) %>% nrow()/n_genes
n_celltype_gene <- vp_sub %>% dplyr::filter(vp_celltype> 0.01) %>% nrow()/n_genes
n_rel <- n_batch_gene/n_celltype_gene
# Mean variance that is explained by the batch effect/celltype
m_batch <- mean(vp_sub$vp_batch, na.rm = TRUE)
m_celltype <- mean(vp_sub$vp_celltype, na.rm = TRUE)
m_rel <- m_batch/m_celltypeScatterplot batch
plot_dev("vp_batch", "vp_batch")## `geom_smooth()` using formula 'y ~ x'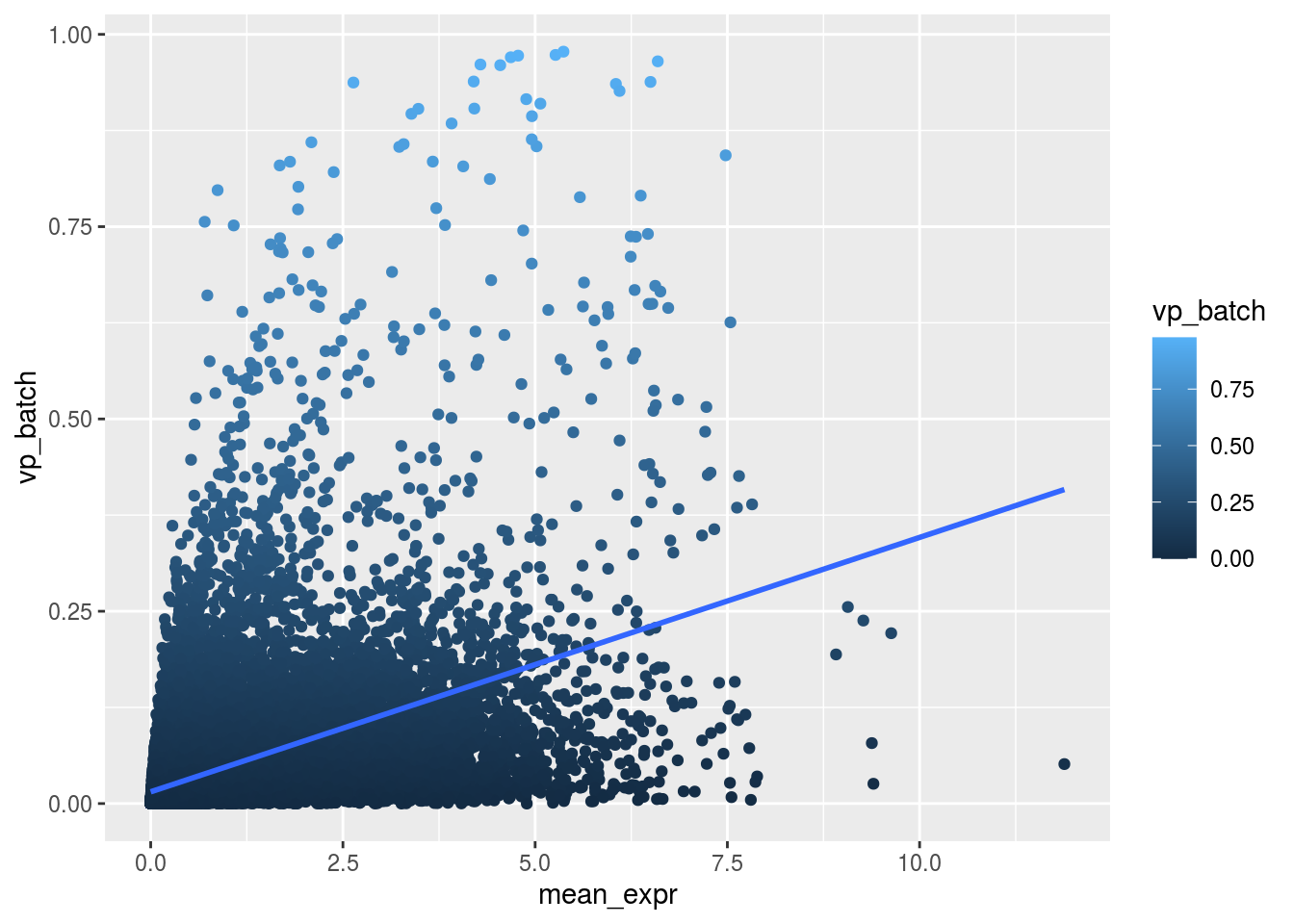
Scatterplot celltype
plot_dev("vp_celltype", "vp_celltype")## `geom_smooth()` using formula 'y ~ x'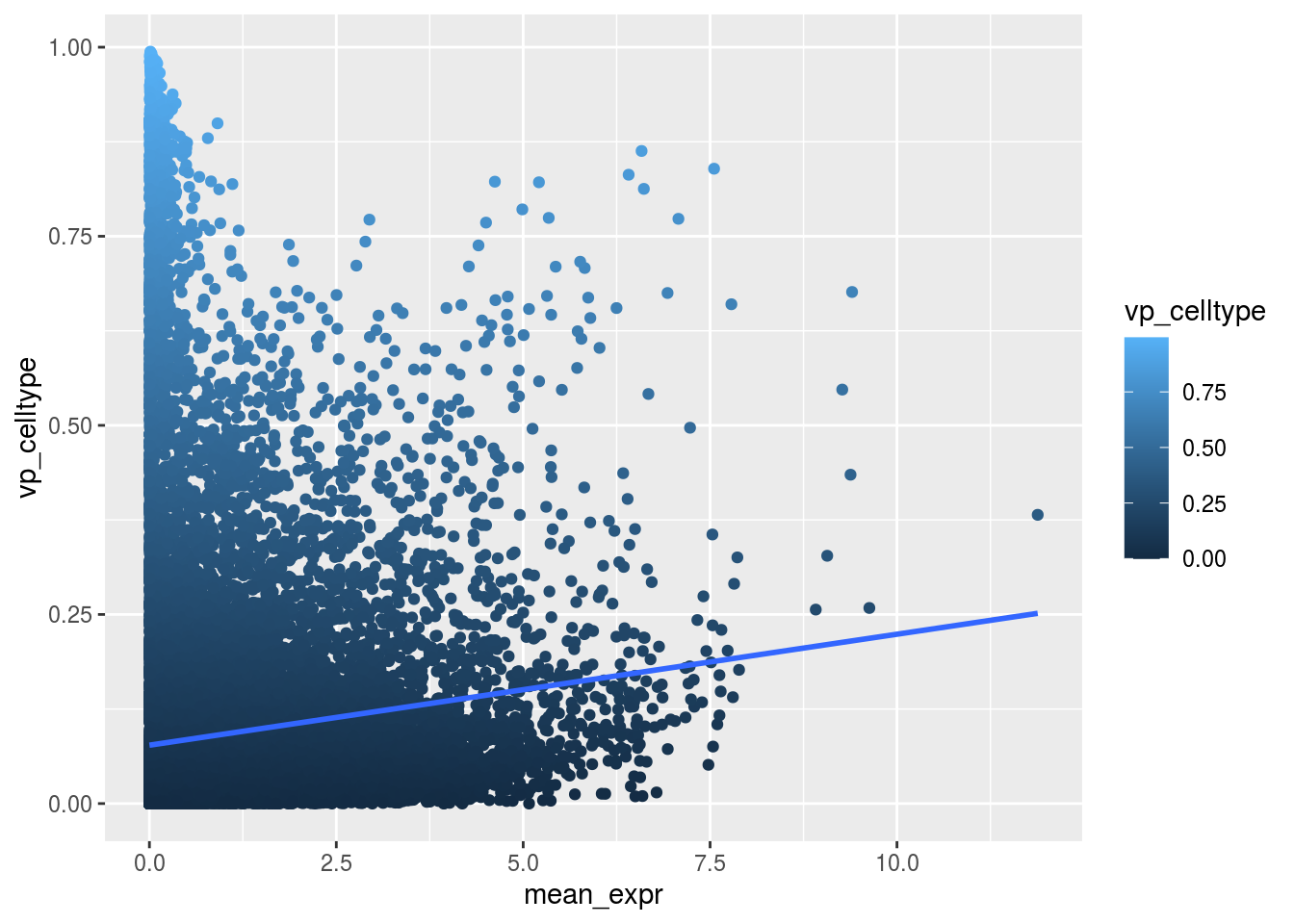
Ternary plot all genes
#t1Ternary plot gene expression classes
#t1 + facet_grid(~expr_class)Cellspecific Mixing score
Overall
#visualize overall cms score
visHist(sce, n_col = 2, prefix = FALSE)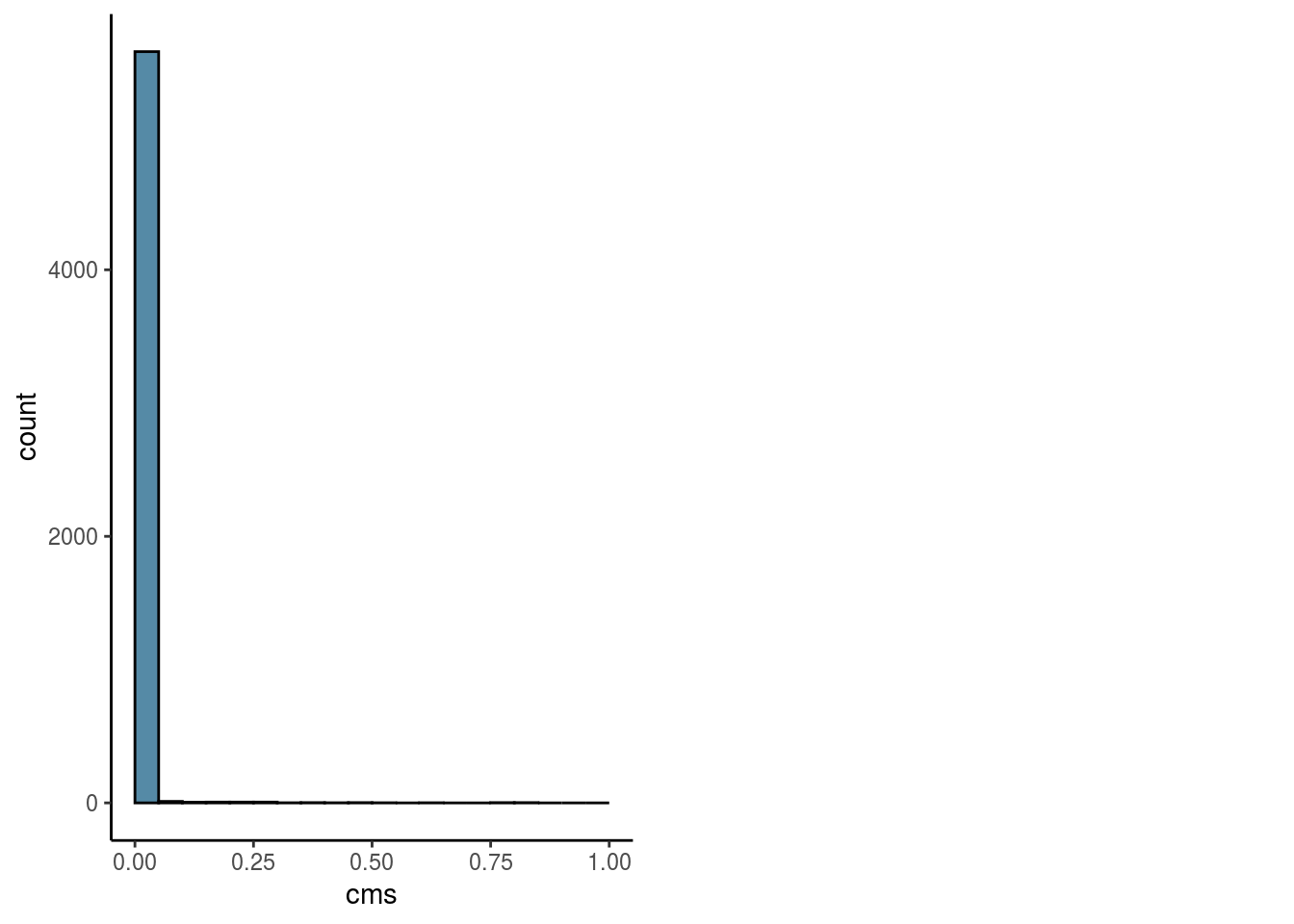
visMetric(sce, metric = "cms_smooth", dim_red = "UMAP")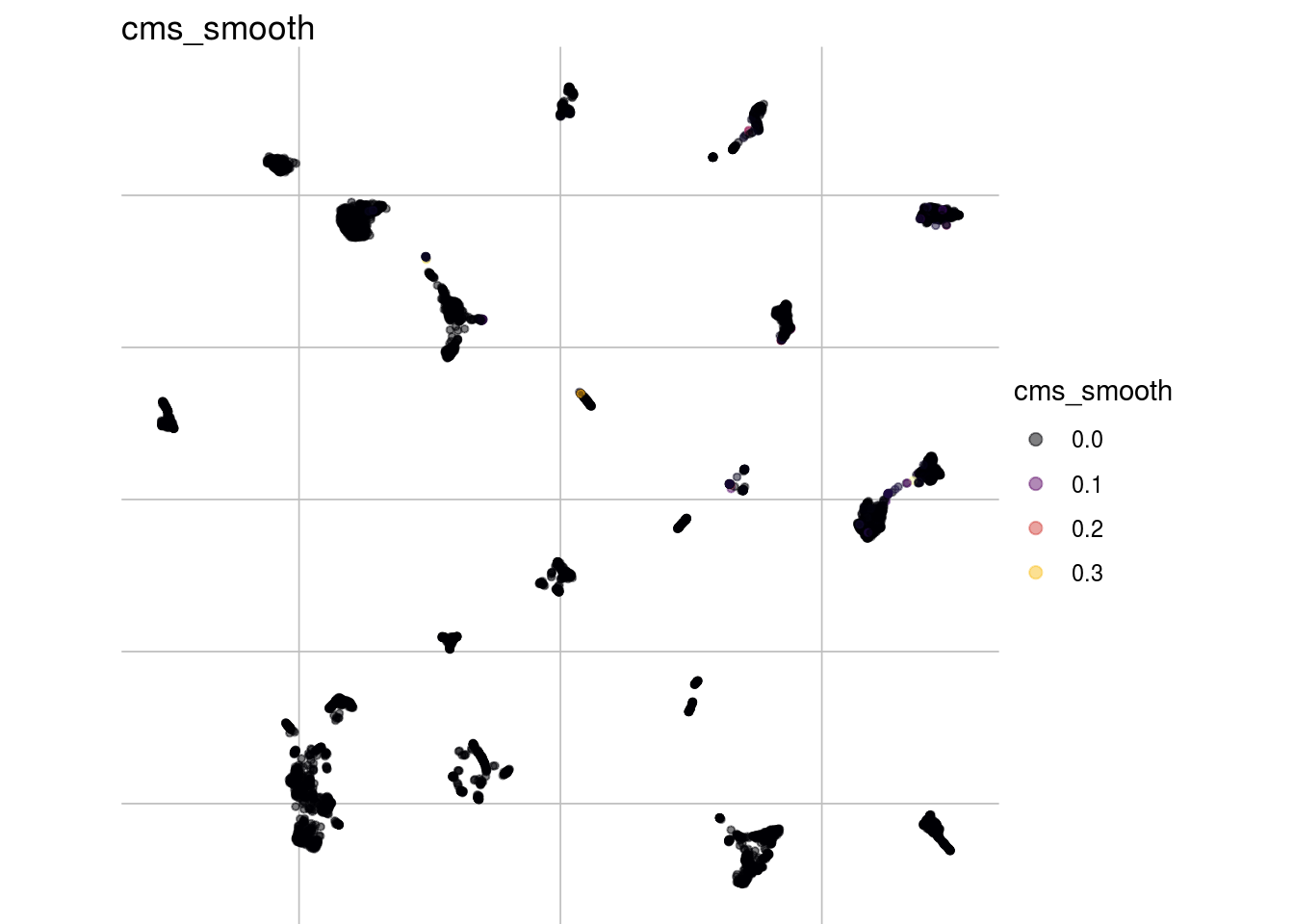
visGroup(sce, celltype, dim_red = "UMAP")
#summarize
mean_cms <- mean(sce$cms)
n_cms_0.01 <- length(which(sce$cms < 0.01))
cluster_mean_cms <- as_tibble(colData(sce)) %>% group_by_at(celltype) %>% summarize(cms_mean = mean(cms))
var_cms <- var(cluster_mean_cms$cms_mean)Celltypes cms smooth
#compare by celltypes
visCluster(sce, metric_var = "cms_smooth", cluster_var = celltype)## Picking joint bandwidth of 0.00136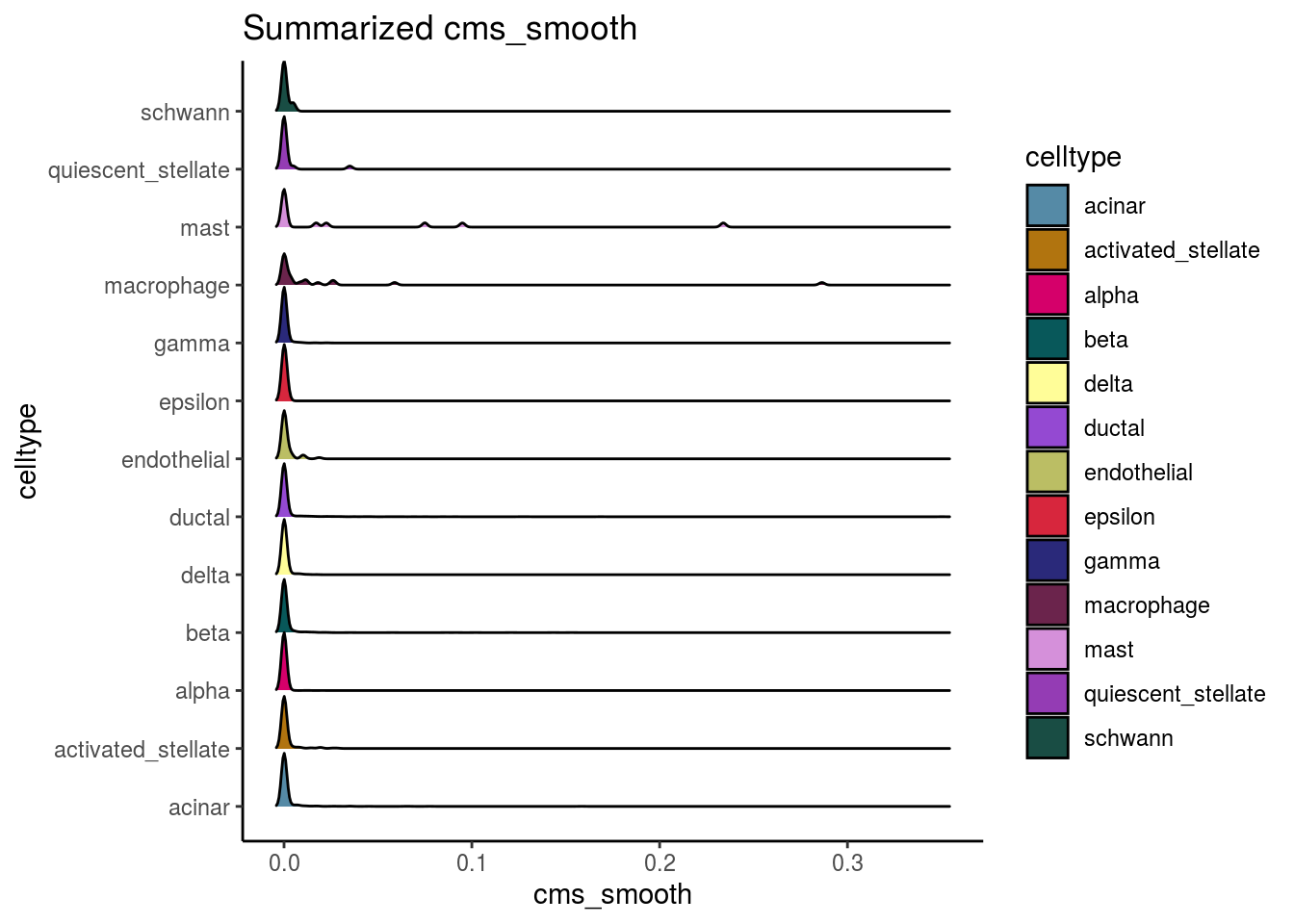
visCluster(sce, metric_var = "cms_smooth", cluster_var = celltype, violin = TRUE)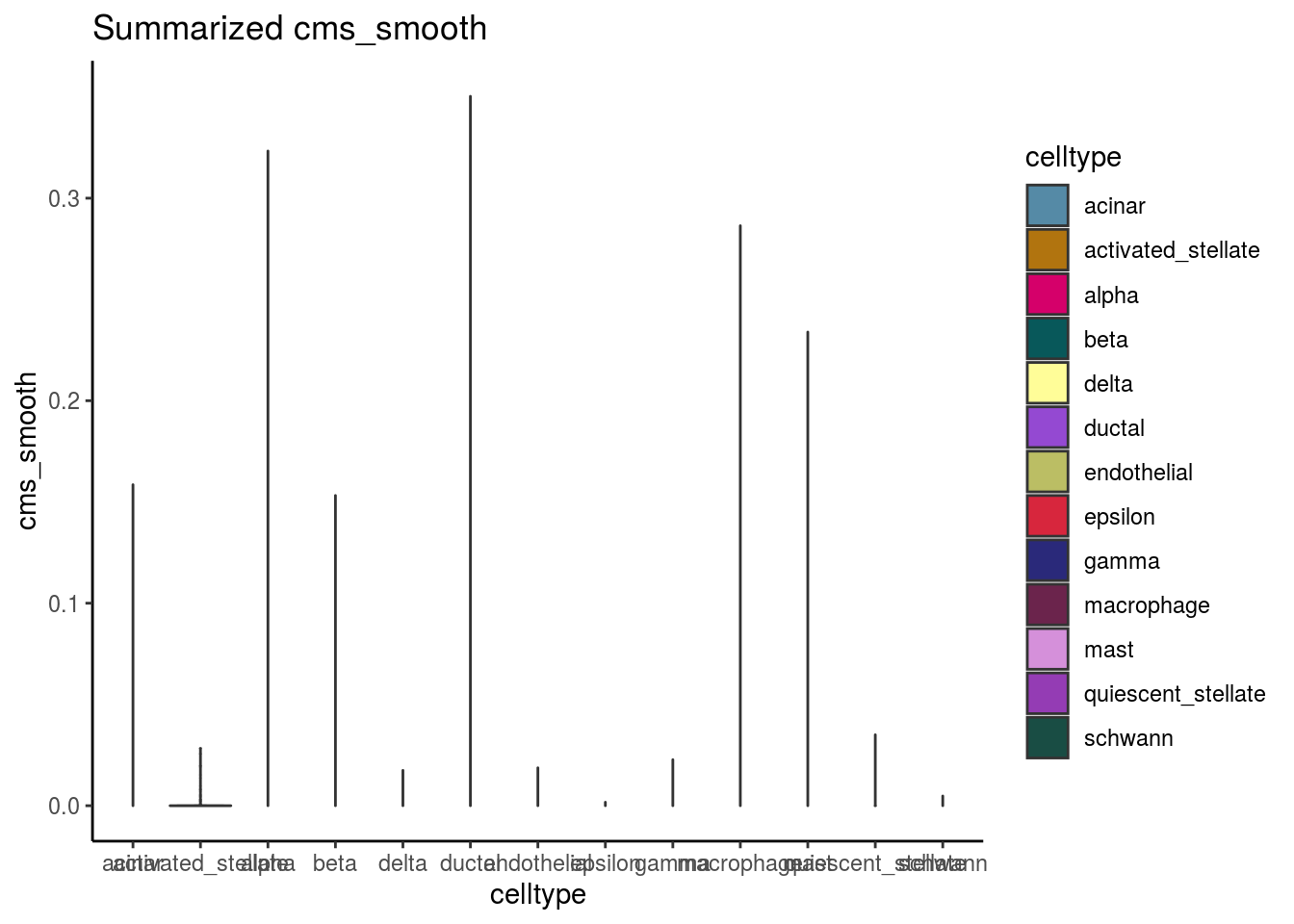
Celltypes histogram
#compare histogram by celltype
p <- ggplot(as.data.frame(colData(sce)),
aes_string(x = "cms", fill = celltype)) +
geom_histogram() +
facet_wrap(celltype, scales = "free_y", ncol = 3) +
scale_fill_manual(values = cols) +
theme_classic()
p + geom_vline(aes_string(xintercept = "cms_mean",
colour = celltype),
cluster_mean_cms, linetype=2) +
scale_color_manual(values = cols) ## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.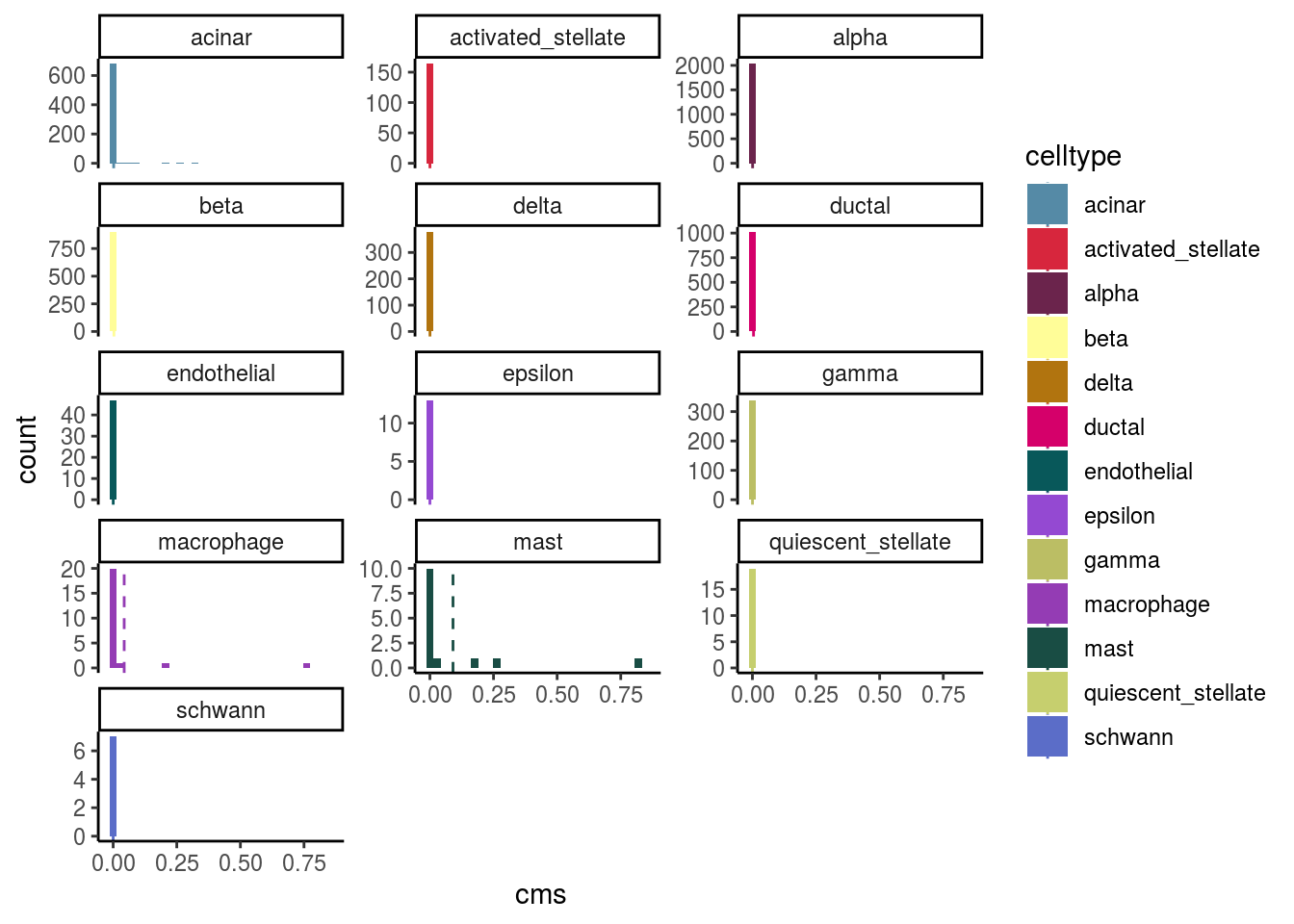
Celltype specificity
Celltype abundance
meta_tib <- as_tibble(colData(sce)) %>% group_by_at(c(batch, celltype)) %>% summarize(n = n()) %>% dplyr::mutate(cell_freq = n / sum(n))
plot_abundance <- function(cluster_var, tib, x_var){
meta_df <- as.data.frame(eval(tib))
p <- ggplot(data=meta_df, aes_string(x=x_var, y="cell_freq", fill = cluster_var)) +
geom_bar(stat="identity") + scale_fill_manual(values=cols, name = "celltype")
p + coord_flip() + theme_minimal()
}
plot_abundance(cluster_var = celltype, tib = meta_tib, x_var = batch)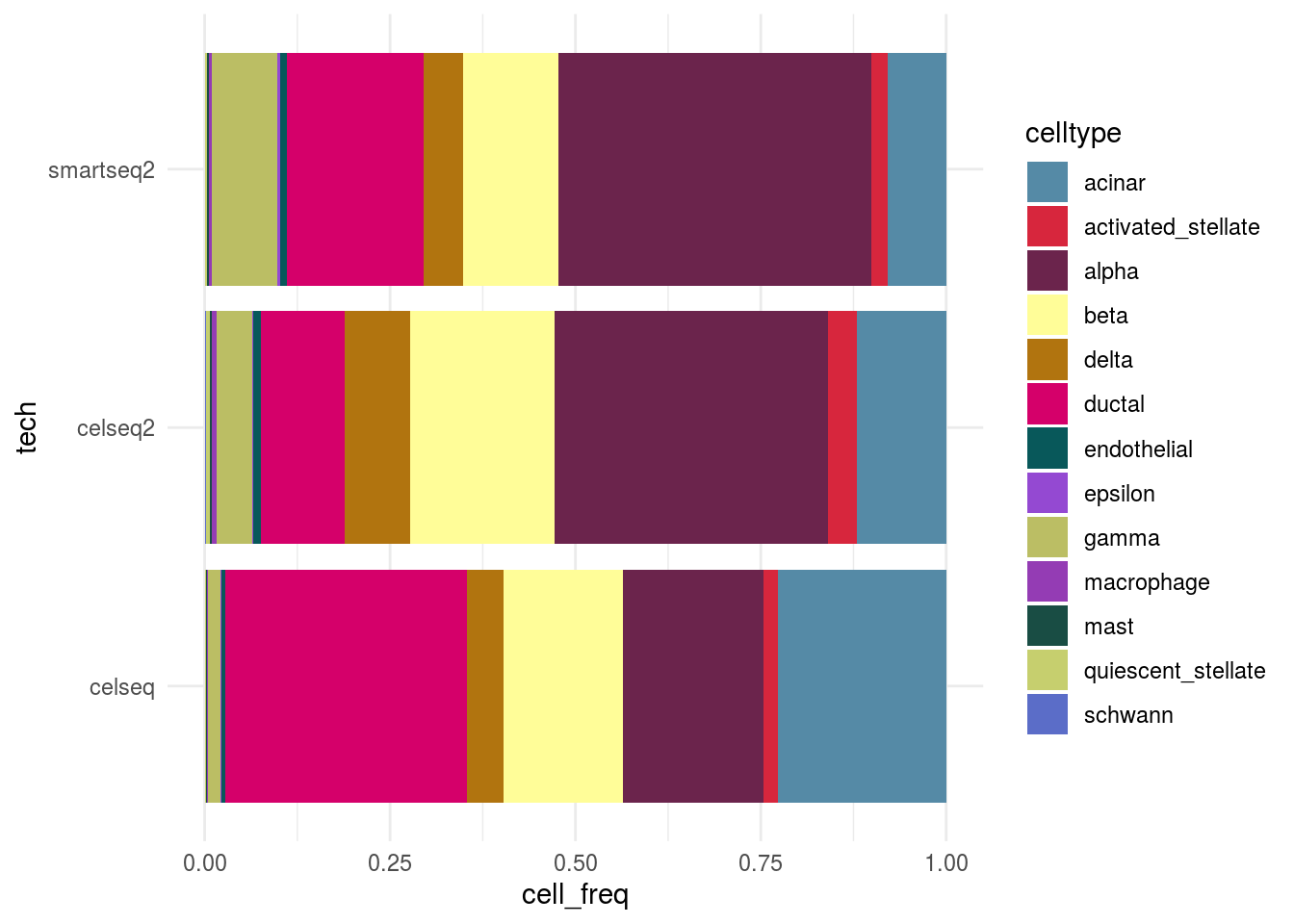
#summarize diff abundance
mean_rel_abund_diff <- mean(unlist(abund))
min_rel_abund_diff <- min(unlist(abund))
max_rel_abund_diff <- max(unlist(abund))Batch and celltype specific count distributions
Do the overall count distribution vary between batches? Are count distributions celltype depended
#batch level
bids <- levels(as.factor(colData(sce)[, batch]))
names(bids) <- bids
cids <- levels(as.factor(colData(sce)[, celltype]))
names(cids) <- cids
#mean gene expression by batch and cluster
mean_list <- lapply(bids, function(batch_var){
mean_cluster <- lapply(cids, function(cluster_var){
counts_sc <- as.matrix(logcounts(
sce[, colData(sce)[, batch] %in% batch_var &
colData(sce)[, celltype] %in% cluster_var]))
})
mean_c <- mean_cluster %>% map(rowMeans) %>% bind_rows %>%
dplyr::mutate(gene=rownames(sce)) %>%
gather(cluster, logcounts, cids)
})## Note: Using an external vector in selections is ambiguous.
## ℹ Use `all_of(cids)` instead of `cids` to silence this message.
## ℹ See <https://tidyselect.r-lib.org/reference/faq-external-vector.html>.
## This message is displayed once per session.mean_expr <- mean_list %>% bind_rows(.id= "batch")
ggplot(mean_expr, aes(x=logcounts, colour=batch)) + geom_density(alpha=.3) +
theme_classic() +
facet_wrap( ~ cluster, ncol = 3) +
scale_colour_manual(values = cols) +
scale_x_continuous(limits = c(0, 7))## Warning: Removed 4071 rows containing non-finite values (stat_density).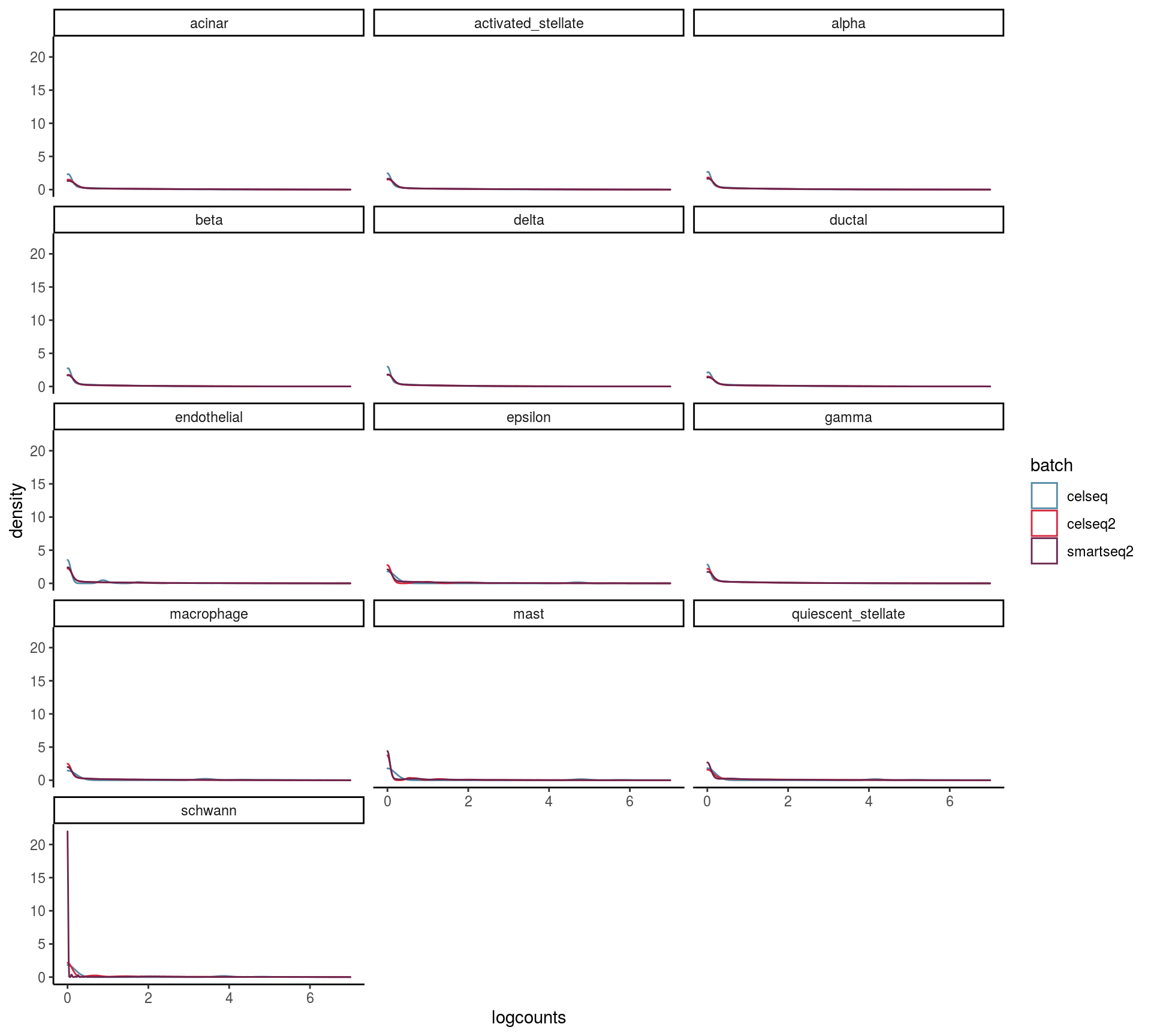
Batch to batch comparisons of expression distributions
Differentially expressed genes
Upset plot
## Upset plot\
cont <- param[["cont"]]
cs <- names(cont)
names(cs) <- cs
# Filter DEG by pvalue
FilterDEGs <- function (degDF = df, filter = c(FDR = 5)){
rownames(degDF) <- degDF$gene
#pval <- degDF[, grep("adj.P.Val$", colnames(degDF)), drop = FALSE]
pval <- degDF[, grep("PValue$", colnames(degDF)), drop = FALSE]
pf <- pval <= filter["FDR"]/100
pf[is.na(pf)] <- FALSE
DEGlistUPorDOWN <- sapply(colnames(pf), function(x) rownames(pf[pf[, x, drop = FALSE], , drop = FALSE]), simplify = FALSE)
}
result <- list()
m2 <- list()
for(jj in 1:length(cs)){
result[[jj]] <- sapply(res_de[[1]][[names(cs)[jj]]], function(x) FilterDEGs(x))
names(result[[jj]]) <- cids
m2[[jj]] = make_comb_mat(result[[jj]], mode = "intersect")
}
names(result) <- names(cs)
names(m2) <- names(cs)
lapply(m2, function(x) UpSet(x))## $`celseq-celseq2`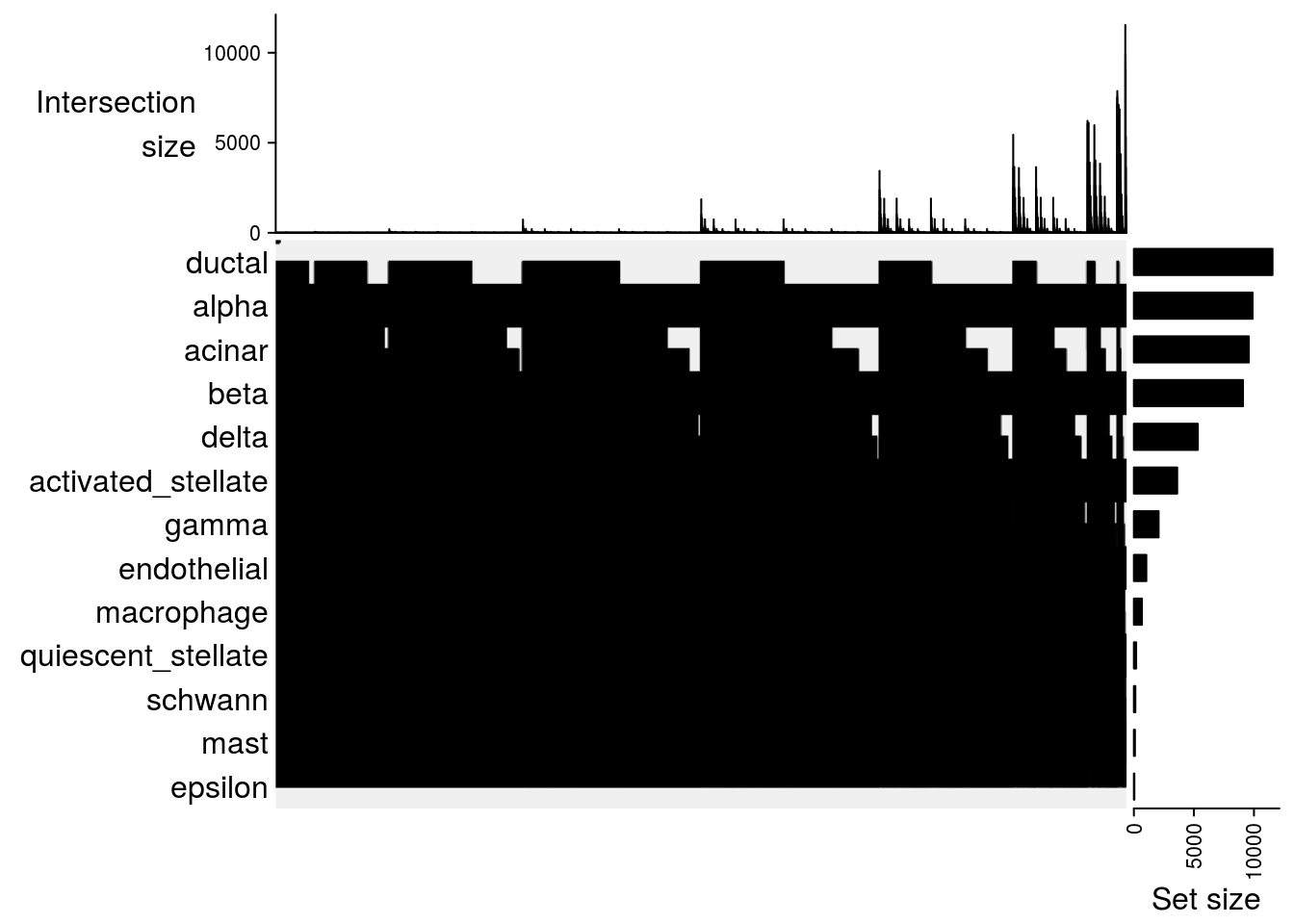
##
## $`celseq-smartseq2`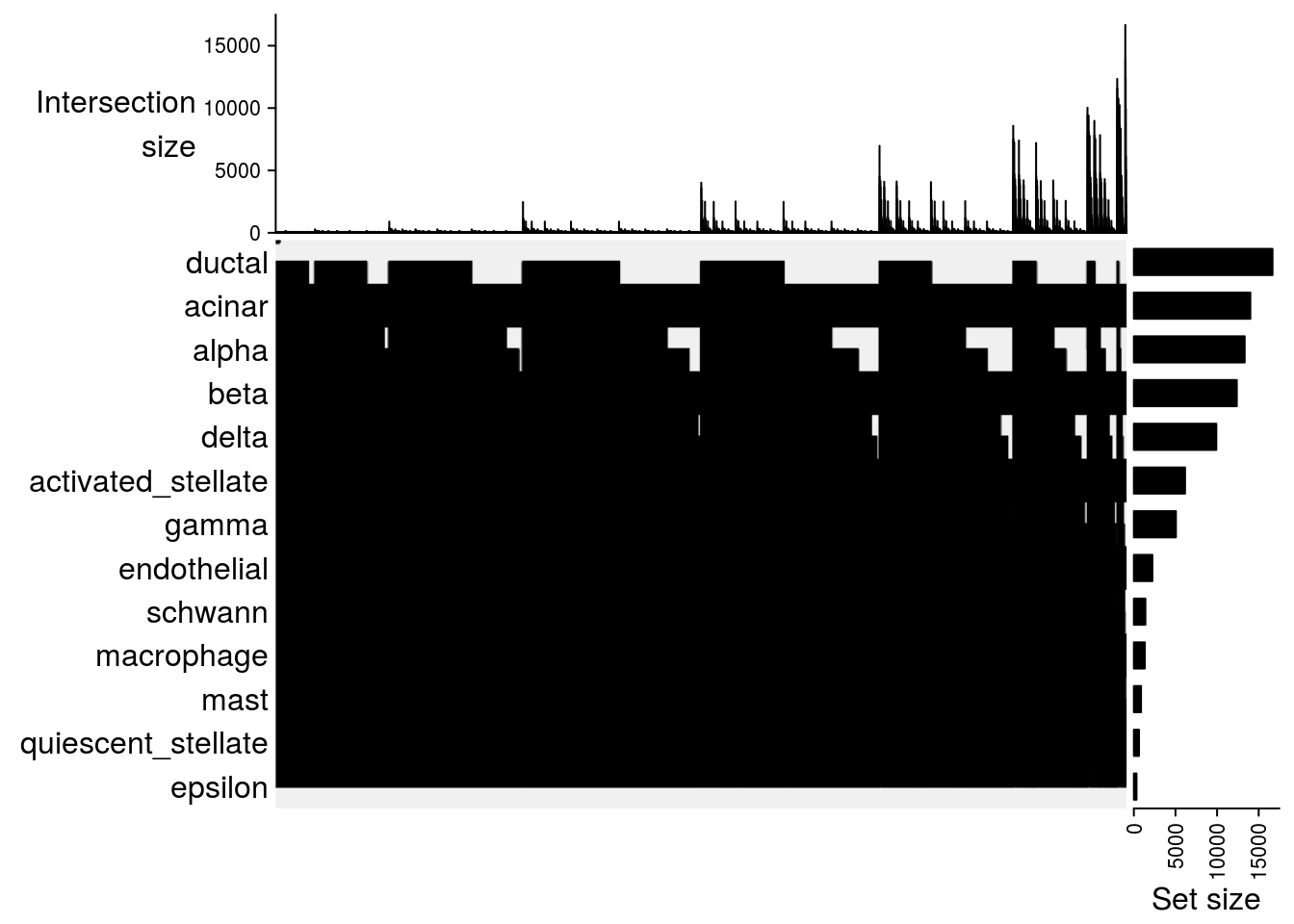
##
## $`celseq2-smartseq2`
Logfold_change and GSEA
# DE genes (per cluster and mean)
res <- res_de[["table"]]
#n_de <- lapply(res, function(y) vapply(y, function(x) sum(x$adj.P.Val < 0.05), numeric(1)))
n_de <- lapply(res, function(y) vapply(y, function(x) sum(x$adj.PValue < 0.05), numeric(1)))
n_genes_lfc1 <- lapply(res, function(y) vapply(y, function(x) sum(abs(x$logFC) > 1), numeric(1)))
mean_n_genes_lfc1 <- mean(unlist(n_genes_lfc1))/n_genes
# plot DE for all comparison and check gene sets
#get geneset
gs <- read_delim(params$gs, delim = "\n", col_names = "cat")## Parsed with column specification:
## cols(
## cat = col_character()
## )cats <- sapply(gs$cat, function(u) strsplit(u, "\t")[[1]][-2],
USE.NAMES = FALSE)
names(cats) <- sapply(cats, .subset, 1)
cats <- lapply(cats, function(u) u[-1])
plotDE <- function(cont_var){
#res_s <- res[[cont_var]] %>% map(filter, adj.P.Val < .05) %>% map(filter, abs(logFC) > 1)
res_s <- res[[cont_var]] %>% map(dplyr::filter, PValue < .05) %>% map(dplyr::filter, abs(logFC) > 1)
#plot
lapply(names(res[[cont_var]]), function(ct){
ct_de <- res[[cont_var]][[ct]]
ct_de$gene <- gsub('[A-z0-9]*\\.', '', ct_de$gene)
res_s[[ct]]$gene <- gsub('[A-z0-9]*\\.', '', res_s[[ct]]$gene)
#p <- ggplot(ct_de, aes(x = AveExpr, y = logFC, colour = abs(logFC) > 1, label = gene)) +
p <- ggplot(ct_de, aes(x = logCPM, y = logFC, colour = abs(logFC) > 2, label = gene)) +
geom_point(size = 2, alpha = .5) +
geom_label_repel(data = res_s[[ct]]) +
ggtitle(paste0(ct,": ", cont_var)) +
theme_classic()
print(p)
cat("Cluster:", ct, "Contrast:", cont_var,
"Num genes:", nrow(ct_de), "Num DE:", nrow(res_s[[ct]]), "\n" )
# run 'camera' for this comparison
inds <- ids2indices(cats, ct_de$gene)
#cm <- cameraPR(ct_de$t, inds) #LR
cm <- cameraPR(ct_de$F, inds)
print(cm %>% rownames_to_column("category") %>%
filter(FDR < .05 & NGenes >= 5) %>% head(8))
})
}
if( length(names(res)) <= 3 ){
pathways <- lapply(names(res), plotDE)
}
## Cluster: acinar Contrast: celseq-celseq2 Num genes: 24509 Num DE: 4348
## category
## 1 GO_RRNA_BINDING
## 2 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 3 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 4 GO_TRANSLATION_REGULATOR_ACTIVITY
## 5 GO_PROTON_TRANSPORTING_ATP_SYNTHASE_ACTIVITY_ROTATIONAL_MECHANISM
## 6 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H
## 7 GO_HYDROGEN_EXPORTING_ATPASE_ACTIVITY
## 8 GO_INSULIN_LIKE_GROWTH_FACTOR_RECEPTOR_BINDING
## NGenes Direction PValue FDR
## 1 57 Up 2.333972e-28 2.102909e-25
## 2 204 Up 1.618215e-24 7.290057e-22
## 3 52 Up 2.135962e-15 6.415006e-13
## 4 34 Up 1.003680e-13 2.260788e-11
## 5 11 Up 8.930456e-11 1.609268e-08
## 6 93 Up 4.822641e-10 7.241999e-08
## 7 27 Up 3.727108e-09 4.797321e-07
## 8 15 Up 9.594308e-09 1.080559e-06
## Cluster: activated_stellate Contrast: celseq-celseq2 Num genes: 24509 Num DE: 3191
## category
## 1 GO_INTRAMOLECULAR_OXIDOREDUCTASE_ACTIVITY_TRANSPOSING_C_C_BONDS
## 2 GO_RRNA_BINDING
## 3 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 4 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 5 GO_CHEMOATTRACTANT_ACTIVITY
## 6 GO_TRANSLATION_REGULATOR_ACTIVITY
## 7 GO_INTRAMOLECULAR_OXIDOREDUCTASE_ACTIVITY
## 8 GO_PROTEIN_PHOSPHATASE_1_BINDING
## NGenes Direction PValue FDR
## 1 13 Up 2.071237e-34 1.866184e-31
## 2 57 Up 5.799069e-27 2.612480e-24
## 3 204 Up 1.547924e-19 4.648932e-17
## 4 52 Up 9.497443e-14 2.139299e-11
## 5 27 Up 2.364263e-13 4.260402e-11
## 6 34 Up 1.067998e-12 1.603778e-10
## 7 53 Up 6.441151e-11 8.290681e-09
## 8 19 Up 1.168321e-10 1.315821e-08
## Cluster: alpha Contrast: celseq-celseq2 Num genes: 24509 Num DE: 5495
## category
## 1 GO_RAGE_RECEPTOR_BINDING
## 2 GO_LONG_CHAIN_FATTY_ACID_BINDING
## 3 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 4 GO_INTRAMOLECULAR_OXIDOREDUCTASE_ACTIVITY_TRANSPOSING_C_C_BONDS
## 5 GO_PROTON_TRANSPORTING_ATP_SYNTHASE_ACTIVITY_ROTATIONAL_MECHANISM
## 6 GO_MHC_CLASS_II_PROTEIN_COMPLEX_BINDING
## 7 GO_FATTY_ACID_BINDING
## 8 GO_HYDROGEN_EXPORTING_ATPASE_ACTIVITY
## NGenes Direction PValue FDR
## 1 11 Up 3.389480e-37 3.053921e-34
## 2 14 Up 5.001663e-15 2.253249e-12
## 3 52 Up 1.059666e-11 3.182531e-09
## 4 13 Up 3.042316e-11 6.852817e-09
## 5 11 Up 3.288138e-09 5.925226e-07
## 6 15 Up 3.815411e-08 5.729475e-06
## 7 30 Up 6.771552e-07 8.715955e-05
## 8 27 Up 1.170801e-06 1.318615e-04
## Cluster: beta Contrast: celseq-celseq2 Num genes: 24509 Num DE: 5952
## category NGenes
## 1 GO_HORMONE_ACTIVITY 115
## 2 GO_RAGE_RECEPTOR_BINDING 11
## 3 GO_INTRAMOLECULAR_OXIDOREDUCTASE_ACTIVITY_TRANSPOSING_C_C_BONDS 13
## 4 GO_RRNA_BINDING 57
## Direction PValue FDR
## 1 Up 1.141494e-22 1.028486e-19
## 2 Up 2.555074e-06 1.151061e-03
## 3 Up 3.760510e-05 1.129407e-02
## 4 Up 1.633769e-04 3.680066e-02
## Cluster: delta Contrast: celseq-celseq2 Num genes: 24509 Num DE: 4539
## category
## 1 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 2 GO_RRNA_BINDING
## 3 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 4 GO_INTRAMOLECULAR_OXIDOREDUCTASE_ACTIVITY_TRANSPOSING_C_C_BONDS
## 5 GO_GLUTATHIONE_PEROXIDASE_ACTIVITY
## 6 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H
## 7 GO_PROTON_TRANSPORTING_ATP_SYNTHASE_ACTIVITY_ROTATIONAL_MECHANISM
## 8 GO_HYDROGEN_EXPORTING_ATPASE_ACTIVITY
## NGenes Direction PValue FDR
## 1 52 Up 2.654651e-13 2.391841e-10
## 2 57 Up 3.189112e-10 1.436695e-07
## 3 204 Up 9.914541e-09 2.977667e-06
## 4 13 Up 8.716430e-08 1.963376e-05
## 5 17 Up 2.796480e-07 5.039257e-05
## 6 93 Up 1.693273e-06 2.542732e-04
## 7 11 Up 4.094945e-06 5.270779e-04
## 8 27 Up 8.749306e-06 9.853906e-04
## Cluster: ductal Contrast: celseq-celseq2 Num genes: 24509 Num DE: 5885
## category
## 1 GO_RRNA_BINDING
## 2 GO_TRANSLATION_REGULATOR_ACTIVITY
## 3 GO_PROTEIN_PHOSPHATASE_1_BINDING
## 4 GO_SUPEROXIDE_GENERATING_NADPH_OXIDASE_ACTIVITY
## 5 GO_INSULIN_LIKE_GROWTH_FACTOR_RECEPTOR_BINDING
## 6 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 7 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 8 GO_PROTEIN_BINDING_INVOLVED_IN_CELL_CELL_ADHESION
## NGenes Direction PValue FDR
## 1 57 Up 2.084462e-08 1.878101e-05
## 2 34 Up 2.993846e-07 1.348727e-04
## 3 19 Up 5.018901e-06 1.507343e-03
## 4 11 Up 1.066602e-05 2.402520e-03
## 5 15 Up 1.680068e-05 2.856204e-03
## 6 204 Up 1.902023e-05 2.856204e-03
## 7 52 Up 2.574791e-05 3.172573e-03
## 8 11 Up 3.007454e-05 3.172573e-03
## Cluster: endothelial Contrast: celseq-celseq2 Num genes: 24509 Num DE: 1032
## category
## 1 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 2 GO_ACTIVIN_BINDING
## 3 GO_NEUROPILIN_BINDING
## 4 GO_TRANSFORMING_GROWTH_FACTOR_BETA_BINDING
## 5 GO_SEMAPHORIN_RECEPTOR_BINDING
## 6 GO_TRANSMEMBRANE_RECEPTOR_PROTEIN_SERINE_THREONINE_KINASE_ACTIVITY
## 7 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H
## 8 GO_CHEMOREPELLENT_ACTIVITY
## NGenes Direction PValue FDR
## 1 52 Up 1.267890e-20 1.142369e-17
## 2 12 Up 2.002952e-16 9.023297e-14
## 3 15 Up 1.707036e-15 5.126799e-13
## 4 16 Up 2.424835e-14 5.461941e-12
## 5 22 Up 4.330357e-12 7.803303e-10
## 6 16 Up 2.852114e-11 4.282924e-09
## 7 93 Up 6.652034e-10 8.562118e-08
## 8 27 Up 1.639944e-09 1.846987e-07
## Cluster: epsilon Contrast: celseq-celseq2 Num genes: 24509 Num DE: 40
## category
## 1 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 2 GO_PROTEASOME_BINDING
## 3 GO_THREONINE_TYPE_PEPTIDASE_ACTIVITY
## 4 GO_RRNA_BINDING
## 5 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H
## 6 GO_POLY_A_RNA_BINDING
## 7 GO_NUCLEOSOME_BINDING
## 8 GO_NUCLEOSOMAL_DNA_BINDING
## NGenes Direction PValue FDR
## 1 52 Up 1.723649e-20 1.553007e-17
## 2 16 Up 9.787818e-15 4.409412e-12
## 3 21 Up 1.362494e-12 4.092024e-10
## 4 57 Up 2.115010e-10 4.764060e-08
## 5 93 Up 2.041103e-09 3.678067e-07
## 6 1149 Up 1.159730e-08 1.741527e-06
## 7 44 Up 2.581408e-08 3.205089e-06
## 8 29 Up 2.845806e-08 3.205089e-06
## Cluster: gamma Contrast: celseq-celseq2 Num genes: 24509 Num DE: 1948
## category
## 1 GO_INTRAMOLECULAR_OXIDOREDUCTASE_ACTIVITY_TRANSPOSING_C_C_BONDS
## 2 GO_CHEMOATTRACTANT_ACTIVITY
## 3 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 4 GO_RRNA_BINDING
## 5 GO_HYDROGEN_EXPORTING_ATPASE_ACTIVITY
## 6 GO_PROTON_TRANSPORTING_ATP_SYNTHASE_ACTIVITY_ROTATIONAL_MECHANISM
## 7 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 8 GO_INTRAMOLECULAR_OXIDOREDUCTASE_ACTIVITY
## NGenes Direction PValue FDR
## 1 13 Up 9.857564e-27 8.881666e-24
## 2 27 Up 3.876038e-15 1.445432e-12
## 3 52 Up 4.812758e-15 1.445432e-12
## 4 57 Up 6.309811e-12 1.421285e-09
## 5 27 Up 3.309639e-10 5.963969e-08
## 6 11 Up 5.400592e-10 8.109889e-08
## 7 204 Up 4.543595e-08 5.848255e-06
## 8 53 Up 5.975486e-08 6.729892e-06
## Cluster: macrophage Contrast: celseq-celseq2 Num genes: 24509 Num DE: 673
## category
## 1 GO_ALCOHOL_DEHYDROGENASE_NADP_ACTIVITY
## 2 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_THE_CH_CH_GROUP_OF_DONORS_NAD_OR_NADP_AS_ACCEPTOR
## 3 GO_ALDO_KETO_REDUCTASE_NADP_ACTIVITY
## 4 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 5 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_PAIRED_DONORS_WITH_INCORPORATION_OR_REDUCTION_OF_MOLECULAR_OXYGEN_NAD_P_H_AS_ONE_DONOR_AND_INCORPORATION_OF_ONE_ATOM_OF_OXYGEN
## 6 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_THE_CH_CH_GROUP_OF_DONORS
## 7 GO_STEROID_DEHYDROGENASE_ACTIVITY
## 8 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H
## NGenes Direction PValue FDR
## 1 16 Up 2.892568e-160 2.606203e-157
## 2 24 Up 1.844577e-123 8.309817e-121
## 3 26 Up 2.265905e-119 6.805267e-117
## 4 52 Up 5.785270e-62 1.303132e-59
## 5 37 Up 3.000597e-59 5.407076e-57
## 6 57 Up 8.729086e-47 1.310818e-44
## 7 27 Up 5.489087e-38 7.065239e-36
## 8 93 Up 3.345477e-32 3.767843e-30
## Cluster: mast Contrast: celseq-celseq2 Num genes: 24509 Num DE: 89
## category
## 1 GO_PROTON_TRANSPORTING_ATP_SYNTHASE_ACTIVITY_ROTATIONAL_MECHANISM
## 2 GO_OPSONIN_BINDING
## 3 GO_HYDROGEN_EXPORTING_ATPASE_ACTIVITY
## 4 GO_GLUTATHIONE_PEROXIDASE_ACTIVITY
## 5 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 6 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 7 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H
## 8 GO_COMPLEMENT_BINDING
## NGenes Direction PValue FDR
## 1 11 Up 6.770140e-24 6.099896e-21
## 2 10 Up 5.830696e-19 2.626728e-16
## 3 27 Up 2.042889e-17 6.135477e-15
## 4 17 Up 6.738274e-12 1.517796e-09
## 5 52 Up 4.402360e-11 7.933052e-09
## 6 204 Up 6.742707e-11 1.012530e-08
## 7 93 Up 2.469646e-10 3.178787e-08
## 8 18 Up 2.371430e-09 2.670823e-07
## Cluster: quiescent_stellate Contrast: celseq-celseq2 Num genes: 24509 Num DE: 191
## category
## 1 GO_ACTIVIN_BINDING
## 2 GO_TRANSFORMING_GROWTH_FACTOR_BETA_BINDING
## 3 GO_TRANSMEMBRANE_RECEPTOR_PROTEIN_SERINE_THREONINE_KINASE_ACTIVITY
## 4 GO_R_SMAD_BINDING
## 5 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 6 GO_SMAD_BINDING
## 7 GO_TRANSLATION_INITIATION_FACTOR_ACTIVITY
## 8 GO_MYOSIN_HEAVY_CHAIN_BINDING
## NGenes Direction PValue FDR
## 1 12 Up 8.833315e-41 7.958817e-38
## 2 16 Up 7.978105e-23 3.594136e-20
## 3 16 Up 7.345404e-21 2.206070e-18
## 4 21 Up 1.686000e-13 3.797716e-11
## 5 52 Up 1.640241e-11 2.955714e-09
## 6 70 Up 1.590879e-09 2.388969e-07
## 7 50 Up 2.031104e-09 2.614321e-07
## 8 12 Up 4.320912e-09 4.866427e-07
## Cluster: schwann Contrast: celseq-celseq2 Num genes: 24509 Num DE: 123
## category
## 1 GO_MHC_CLASS_II_PROTEIN_COMPLEX_BINDING
## 2 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 3 GO_PEPTIDE_ANTIGEN_BINDING
## 4 GO_CALCIUM_DEPENDENT_PROTEIN_BINDING
## 5 GO_SNRNA_BINDING
## 6 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 7 GO_MHC_PROTEIN_COMPLEX_BINDING
## 8 GO_GROWTH_FACTOR_BINDING
## NGenes Direction PValue FDR
## 1 15 Up 2.379809e-09 2.144208e-06
## 2 52 Up 5.387574e-09 2.427102e-06
## 3 27 Up 5.197341e-08 1.241915e-05
## 4 59 Up 7.028580e-08 1.241915e-05
## 5 34 Up 7.905058e-08 1.241915e-05
## 6 204 Up 8.270247e-08 1.241915e-05
## 7 18 Up 1.103502e-07 1.420365e-05
## 8 122 Up 1.666193e-07 1.876550e-05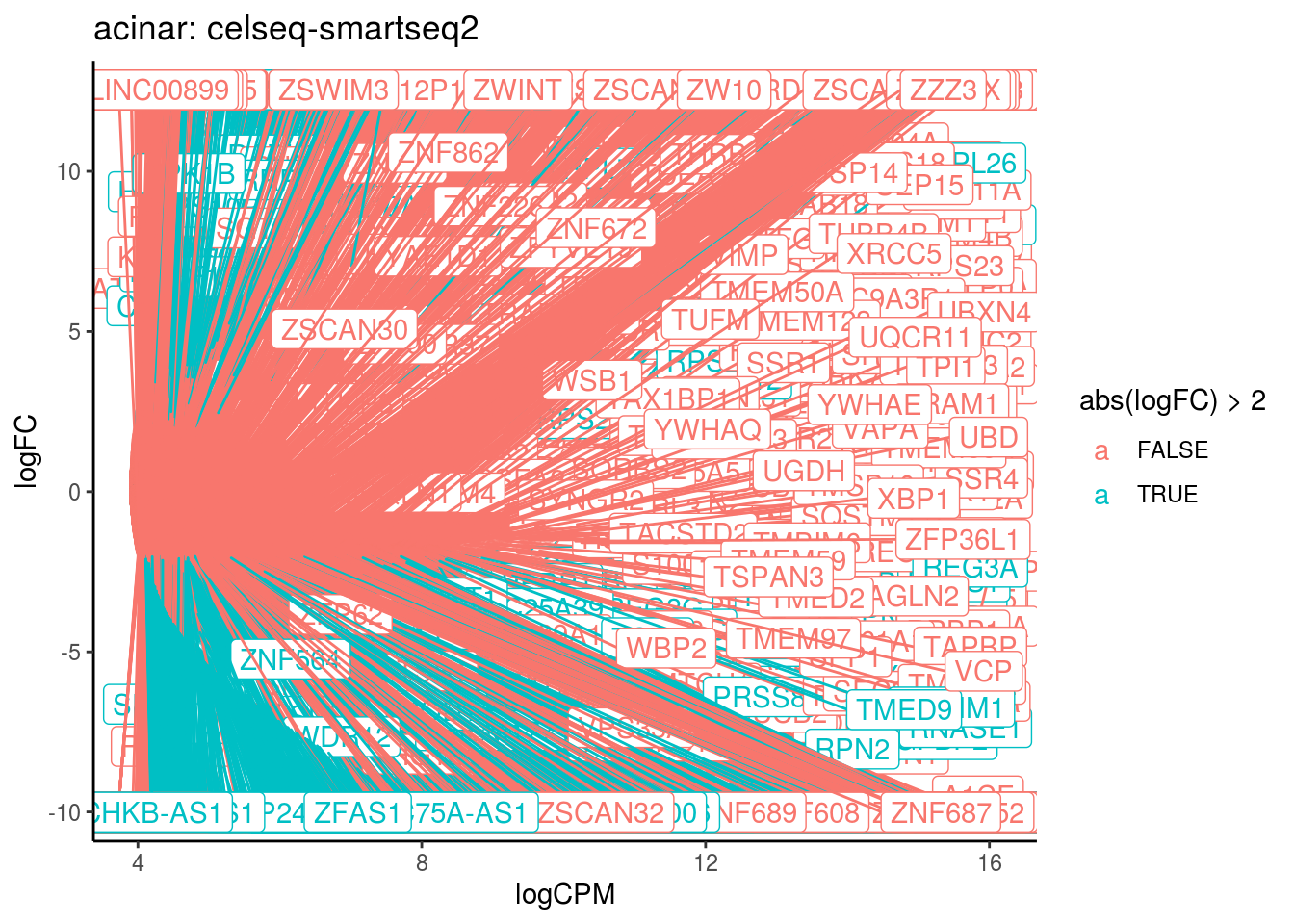
## Cluster: acinar Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 9056
## category NGenes Direction
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up
## 2 GO_POLY_A_BINDING 13 Up
## 3 GO_STRUCTURAL_MOLECULE_ACTIVITY 703 Up
## 4 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_ 18 Up
## PValue FDR
## 1 4.711830e-34 4.245359e-31
## 2 1.084865e-05 4.887317e-03
## 3 2.792452e-05 8.386663e-03
## 4 1.277444e-04 2.877444e-02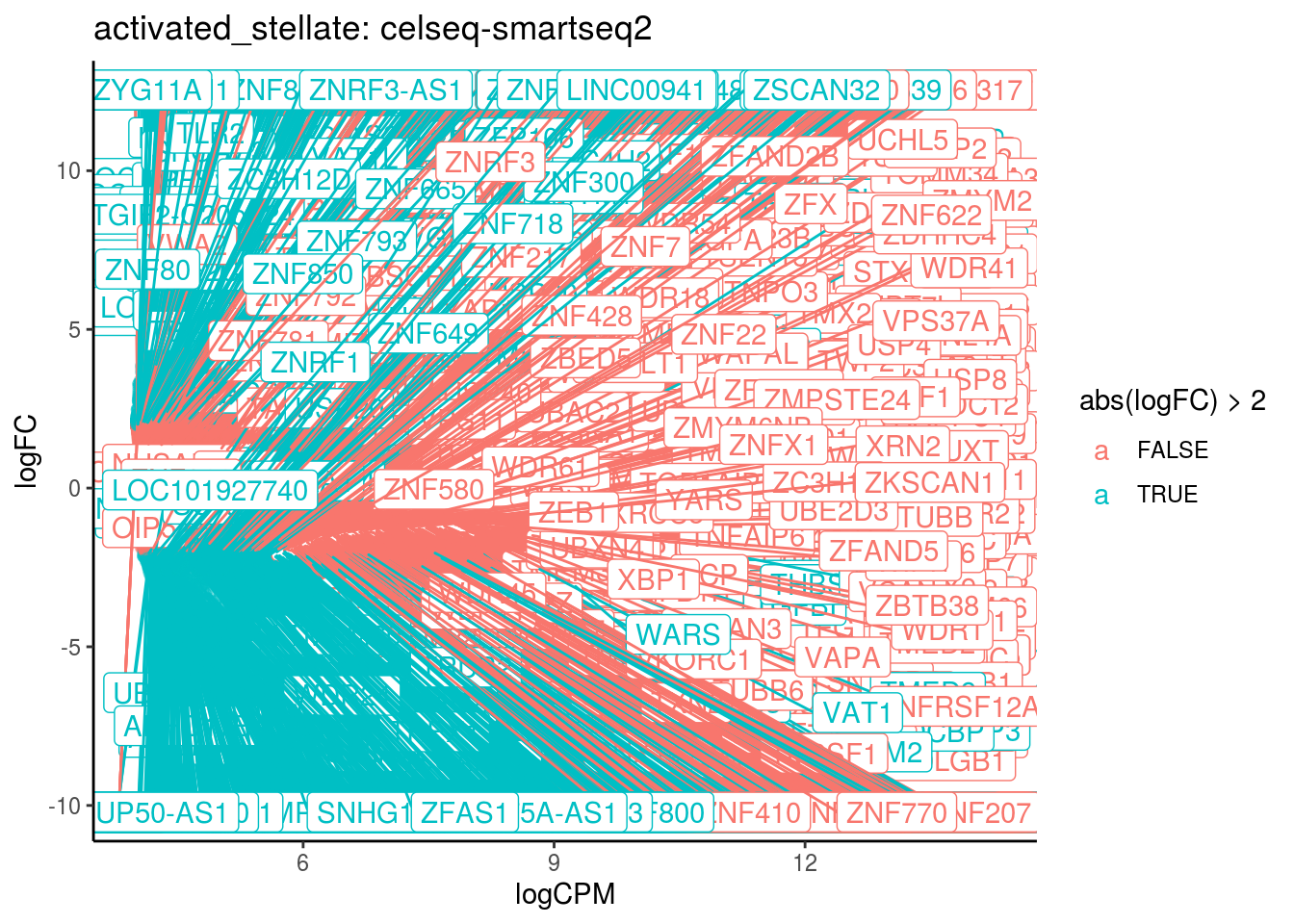
## Cluster: activated_stellate Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 5796
## category NGenes Direction PValue
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up 3.809930e-22
## 2 GO_POLY_A_BINDING 13 Up 6.384585e-09
## 3 GO_POLY_PURINE_TRACT_BINDING 19 Up 5.625977e-06
## FDR
## 1 3.432747e-19
## 2 2.876256e-06
## 3 1.689669e-03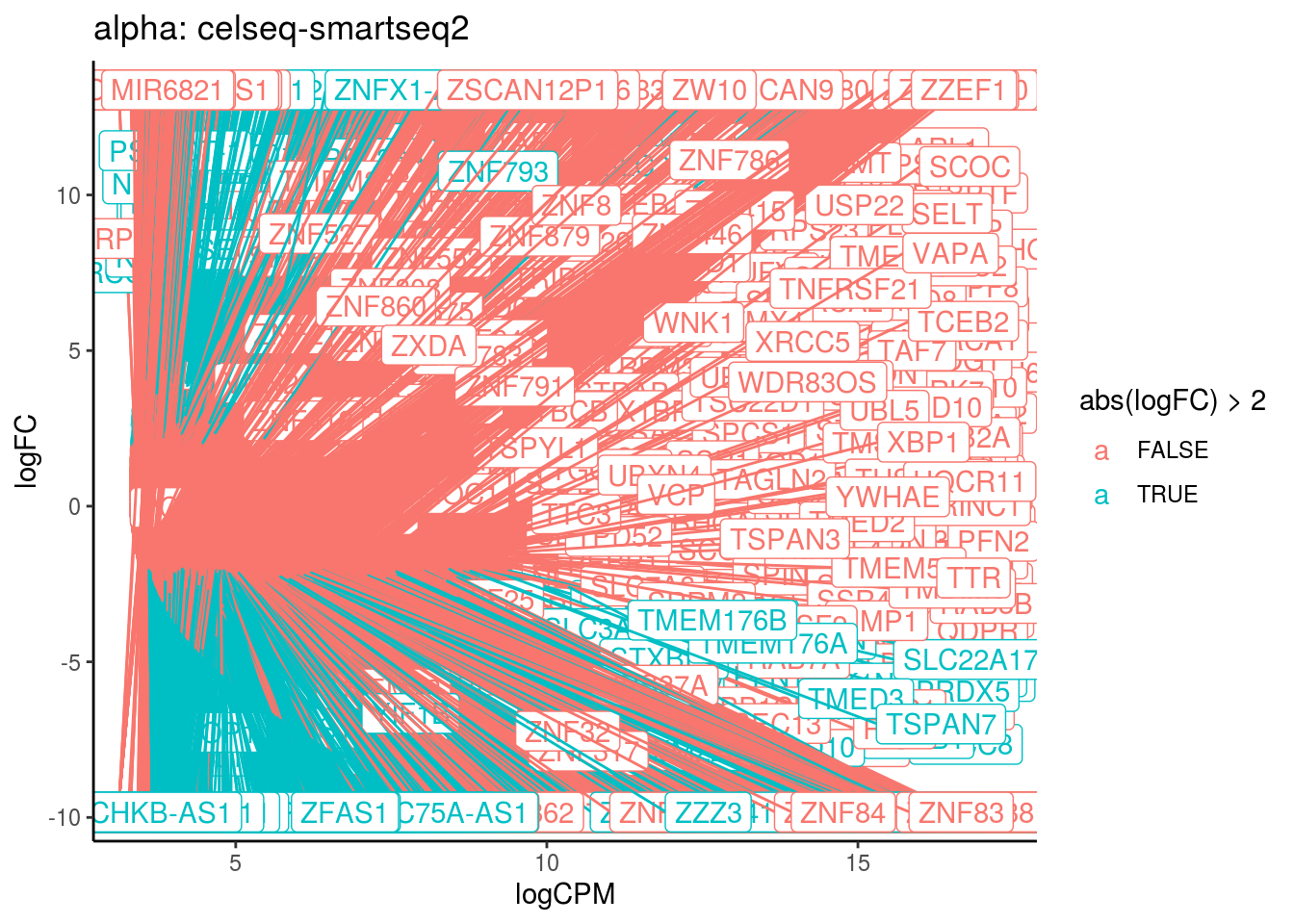
## Cluster: alpha Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 9754
## category NGenes Direction PValue
## 1 GO_POLY_A_BINDING 13 Up 7.710257e-07
## 2 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up 4.172346e-05
## 3 GO_POLY_PURINE_TRACT_BINDING 19 Up 1.273474e-04
## FDR
## 1 0.0006946942
## 2 0.0187964171
## 3 0.0382466658
## Cluster: beta Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 9266
## category NGenes Direction PValue
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up 1.803375e-13
## FDR
## 1 1.624841e-10
## Cluster: delta Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 9417
## category NGenes Direction PValue
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up 1.628133e-07
## FDR
## 1 0.0001466948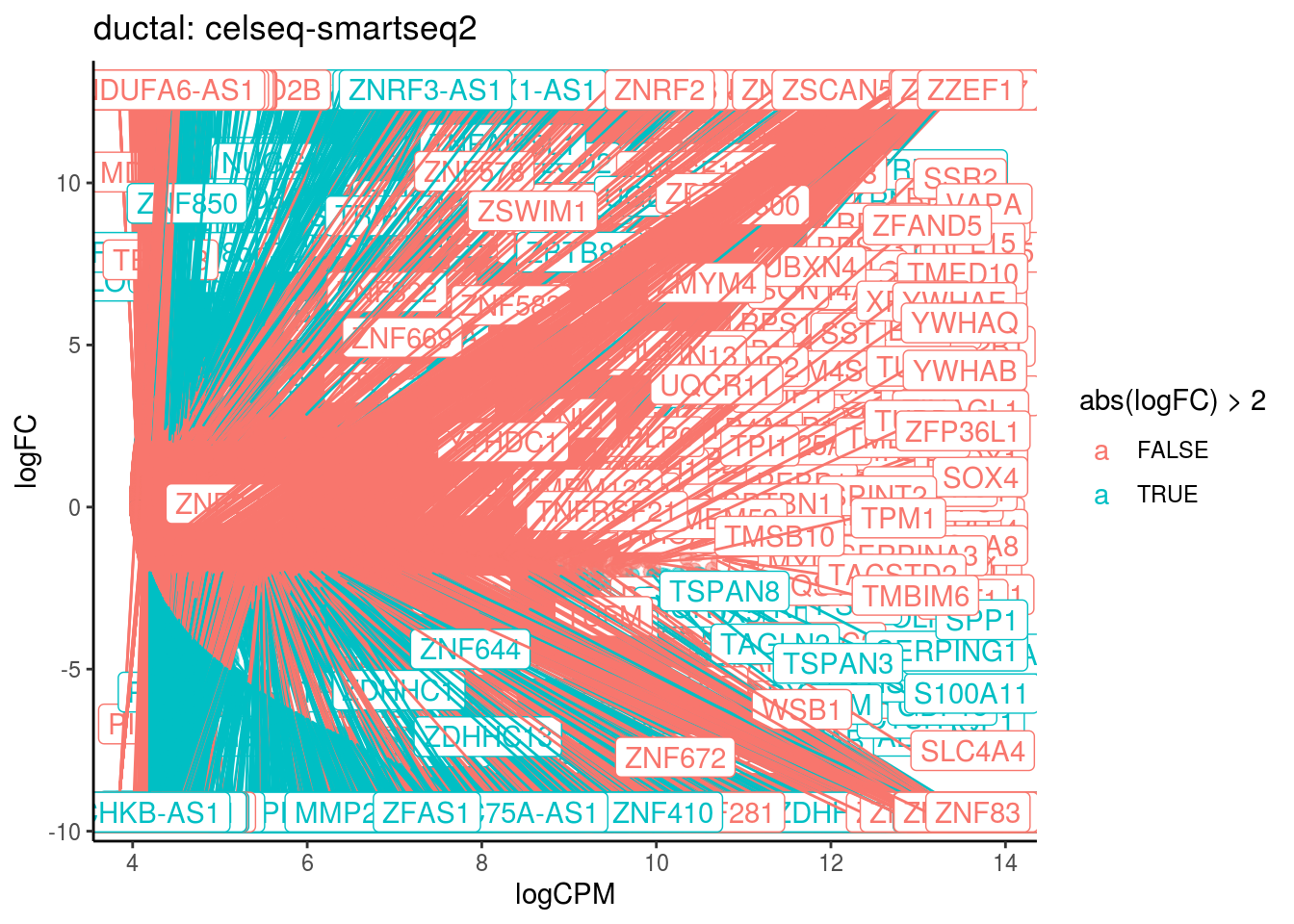
## Cluster: ductal Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 10500
## category NGenes Direction PValue
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up 1.128377e-11
## 2 GO_POLY_A_BINDING 13 Up 1.315098e-05
## FDR
## 1 1.016668e-08
## 2 5.924517e-03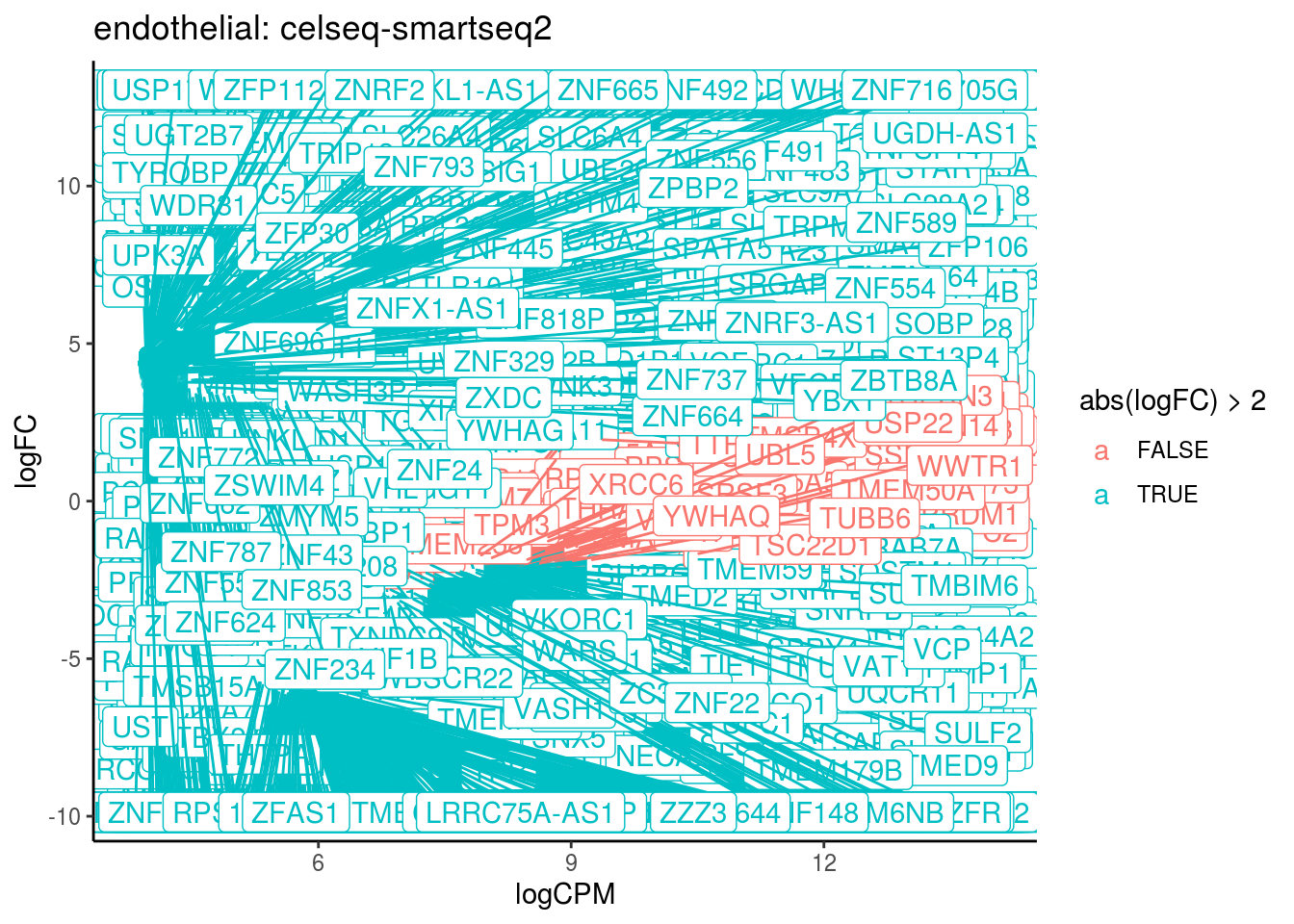
## Cluster: endothelial Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 2220
## category NGenes Direction PValue
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up 1.667636e-07
## FDR
## 1 0.000150254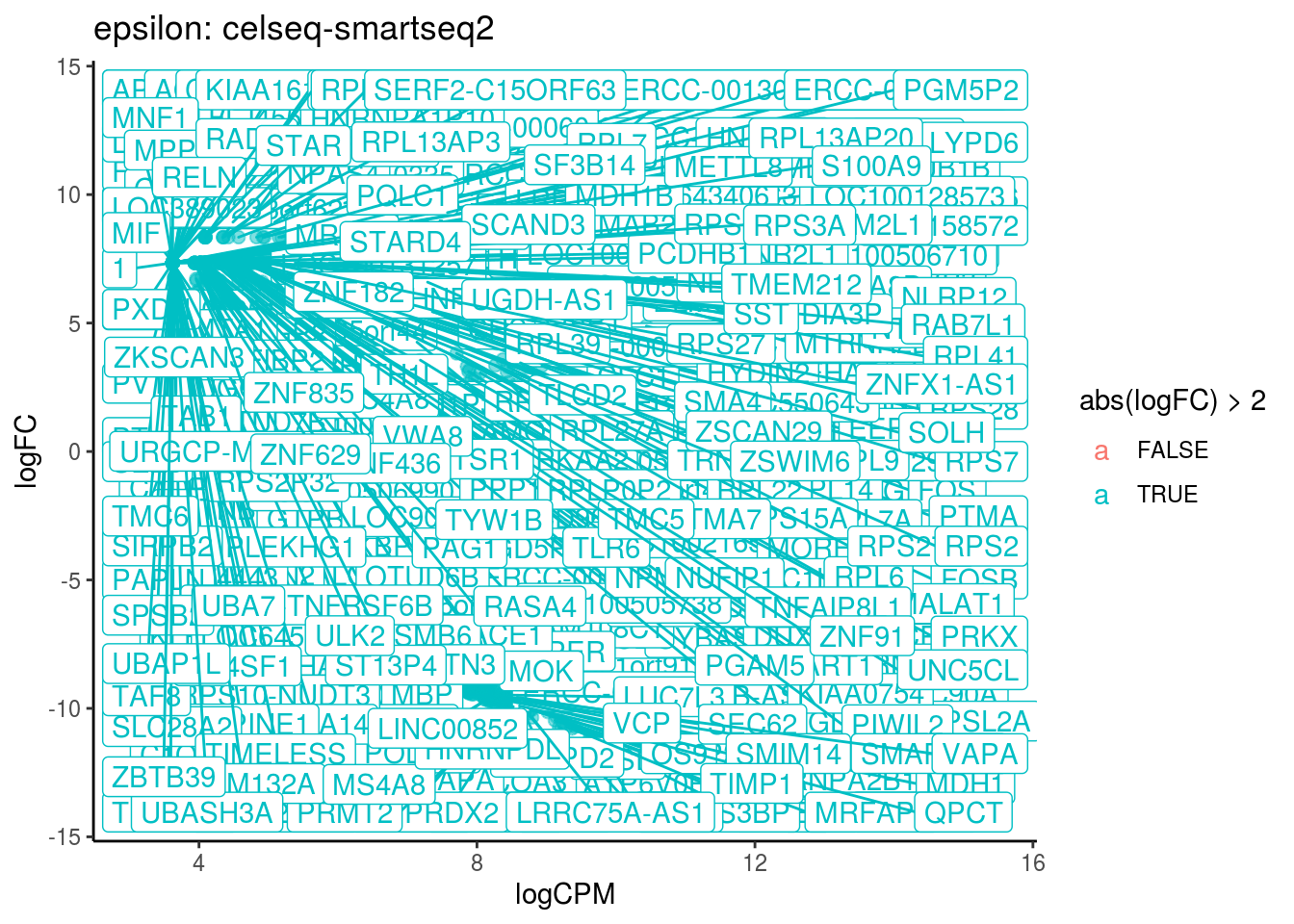
## Cluster: epsilon Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 317
## category NGenes Direction PValue
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up 4.960588e-12
## 2 GO_POLY_A_BINDING 13 Up 2.163754e-05
## FDR
## 1 4.469490e-09
## 2 9.747712e-03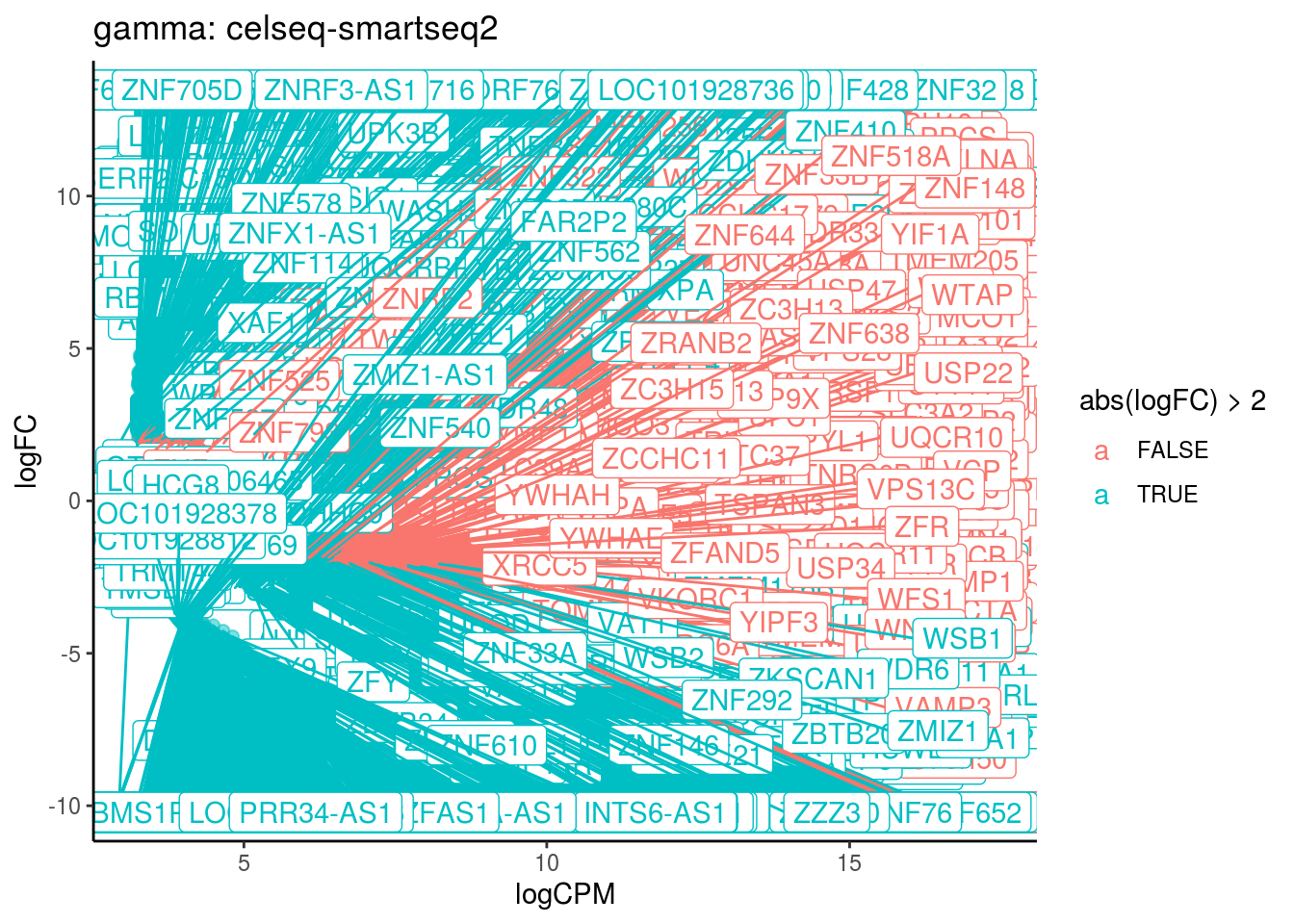
## Cluster: gamma Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 4955
## [1] category NGenes Direction PValue FDR
## <0 rows> (or 0-length row.names)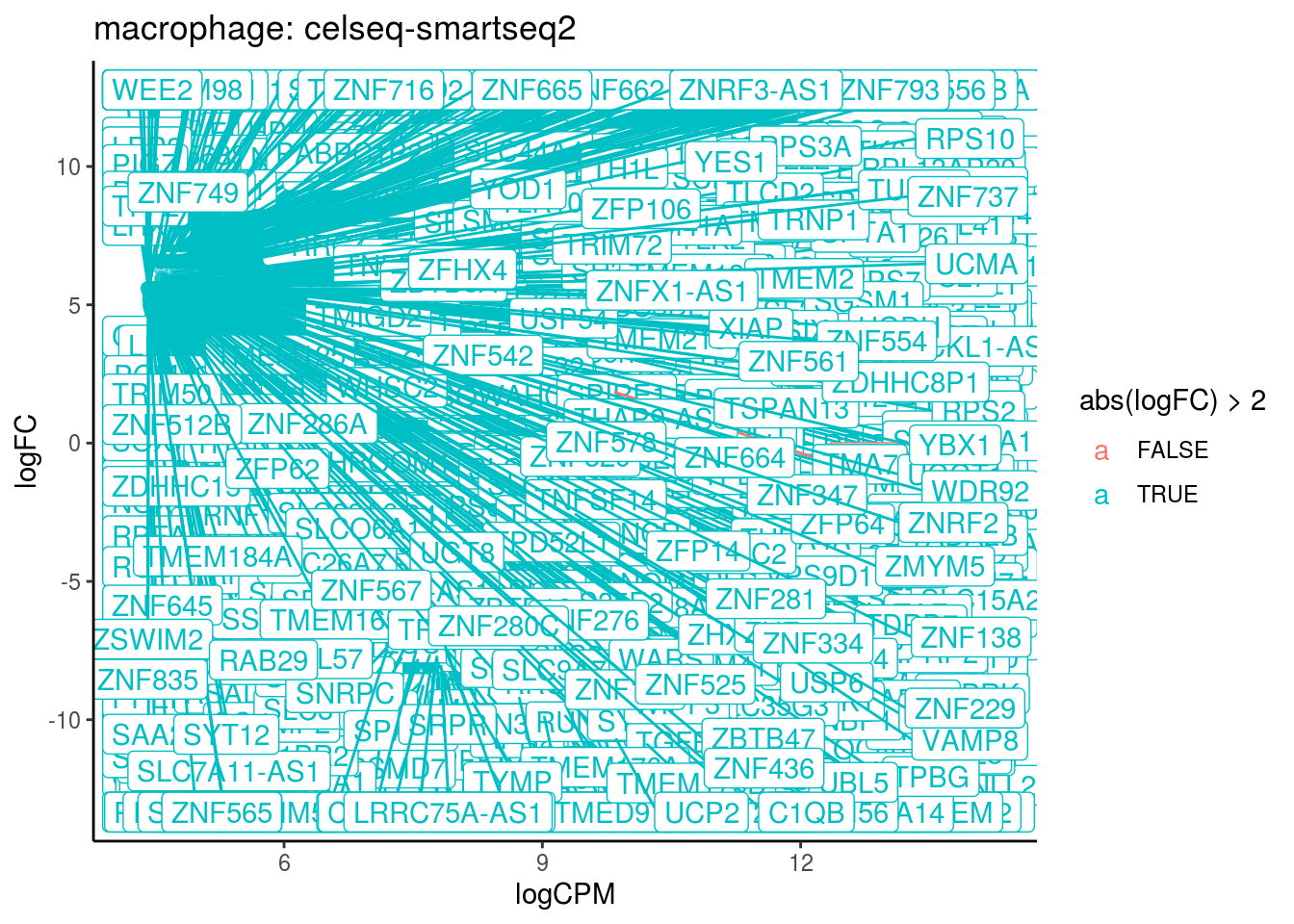
## Cluster: macrophage Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 1315
## category
## 1 GO_ALCOHOL_DEHYDROGENASE_NADP_ACTIVITY
## 2 GO_ALDO_KETO_REDUCTASE_NADP_ACTIVITY
## 3 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_THE_CH_CH_GROUP_OF_DONORS_NAD_OR_NADP_AS_ACCEPTOR
## 4 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_PAIRED_DONORS_WITH_INCORPORATION_OR_REDUCTION_OF_MOLECULAR_OXYGEN_NAD_P_H_AS_ONE_DONOR_AND_INCORPORATION_OF_ONE_ATOM_OF_OXYGEN
## 5 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 6 GO_STEROID_DEHYDROGENASE_ACTIVITY
## 7 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_THE_CH_CH_GROUP_OF_DONORS
## 8 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## NGenes Direction PValue FDR
## 1 16 Up 7.361483e-42 6.632696e-39
## 2 26 Up 1.074886e-31 4.842362e-29
## 3 24 Up 2.010429e-31 6.037988e-29
## 4 37 Up 3.583485e-15 8.071800e-13
## 5 52 Up 2.086470e-11 3.759819e-09
## 6 27 Up 2.944577e-11 4.421773e-09
## 7 57 Up 5.744995e-11 7.394629e-09
## 8 204 Up 3.153479e-10 3.551606e-08
## Cluster: mast Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 860
## category NGenes Direction
## 1 GO_OPSONIN_BINDING 10 Up
## 2 GO_COMPLEMENT_BINDING 18 Up
## 3 GO_LOW_DENSITY_LIPOPROTEIN_PARTICLE_BINDING 15 Up
## 4 GO_PROTEIN_TYROSINE_KINASE_ACTIVATOR_ACTIVITY 15 Up
## 5 GO_LOW_DENSITY_LIPOPROTEIN_PARTICLE_RECEPTOR_BINDING 17 Up
## 6 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up
## 7 GO_SERINE_TYPE_ENDOPEPTIDASE_INHIBITOR_ACTIVITY 89 Up
## 8 GO_POLY_A_BINDING 13 Up
## PValue FDR
## 1 1.900354e-19 1.712219e-16
## 2 1.129847e-14 5.089959e-12
## 3 3.999732e-07 1.201253e-04
## 4 1.463377e-06 3.296256e-04
## 5 1.141417e-05 2.056833e-03
## 6 1.700504e-05 2.553590e-03
## 7 7.022768e-05 8.423288e-03
## 8 7.479057e-05 8.423288e-03
## Cluster: quiescent_stellate Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 613
## category NGenes Direction PValue
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up 7.107612e-13
## 2 GO_R_SMAD_BINDING 21 Up 4.345393e-09
## 3 GO_POLY_A_BINDING 13 Up 9.988803e-08
## 4 GO_CAMP_RESPONSE_ELEMENT_BINDING 13 Up 4.539049e-07
## 5 GO_SMAD_BINDING 70 Up 2.328992e-06
## 6 GO_POLY_PURINE_TRACT_BINDING 19 Up 1.363703e-05
## 7 GO_I_SMAD_BINDING 11 Up 1.609358e-05
## 8 GO_HMG_BOX_DOMAIN_BINDING 17 Up 1.929575e-05
## FDR
## 1 6.403958e-10
## 2 1.957600e-06
## 3 2.999970e-05
## 4 1.022421e-04
## 5 4.196843e-04
## 6 2.047827e-03
## 7 2.071473e-03
## 8 2.173184e-03
## Cluster: schwann Contrast: celseq-smartseq2 Num genes: 24509 Num DE: 1384
## category NGenes Direction PValue
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up 5.054866e-43
## 2 GO_RRNA_BINDING 57 Up 1.625906e-11
## 3 GO_POLY_A_BINDING 13 Up 6.619852e-11
## 4 GO_POLY_A_RNA_BINDING 1149 Up 3.913229e-09
## 5 GO_POLY_PURINE_TRACT_BINDING 19 Up 1.023622e-07
## 6 GO_RNA_BINDING 1549 Up 2.472557e-07
## 7 GO_MRNA_BINDING 152 Up 4.488390e-07
## 8 GO_STRUCTURAL_MOLECULE_ACTIVITY 703 Up 9.779281e-07
## FDR
## 1 4.554434e-40
## 2 7.324706e-09
## 3 1.988162e-08
## 4 8.814549e-07
## 5 1.844568e-05
## 6 3.712956e-05
## 7 5.777199e-05
## 8 1.101391e-04
## Cluster: acinar Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 6586
## category
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 2 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_
## 3 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 4 GO_TRANSLATION_ELONGATION_FACTOR_ACTIVITY
## 5 GO_POLY_A_BINDING
## 6 GO_HYDROGEN_EXPORTING_ATPASE_ACTIVITY
## 7 GO_SINGLE_STRANDED_RNA_BINDING
## 8 GO_PROTON_TRANSPORTING_ATP_SYNTHASE_ACTIVITY_ROTATIONAL_MECHANISM
## NGenes Direction PValue FDR
## 1 204 Up 2.675079e-39 2.410246e-36
## 2 18 Up 6.718721e-12 3.026784e-09
## 3 52 Up 1.654589e-08 4.969282e-06
## 4 19 Up 6.359529e-08 1.432484e-05
## 5 13 Up 1.201213e-07 2.164585e-05
## 6 27 Up 3.333112e-06 4.666300e-04
## 7 68 Up 3.625316e-06 4.666300e-04
## 8 11 Up 4.884584e-06 5.501263e-04
## Cluster: activated_stellate Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 7159
## category NGenes Direction
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up
## 2 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_ 18 Up
## 3 GO_POLY_A_BINDING 13 Up
## 4 GO_POLY_PURINE_TRACT_BINDING 19 Up
## 5 GO_TRANSLATION_ELONGATION_FACTOR_ACTIVITY 19 Up
## 6 GO_SINGLE_STRANDED_RNA_BINDING 68 Up
## 7 GO_HISTONE_LYSINE_N_METHYLTRANSFERASE_ACTIVITY 45 Up
## 8 GO_SINGLE_STRANDED_DNA_BINDING 86 Up
## PValue FDR
## 1 1.591325e-28 1.433784e-25
## 2 1.210974e-18 5.455439e-16
## 3 2.716237e-13 8.157766e-11
## 4 1.196474e-08 2.695058e-06
## 5 7.461787e-08 1.344614e-05
## 6 2.033824e-07 3.054126e-05
## 7 6.107523e-06 7.861255e-04
## 8 1.033130e-05 1.163563e-03
## Cluster: alpha Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 6855
## category NGenes Direction
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up
## 2 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_ 18 Up
## 3 GO_POLY_A_BINDING 13 Up
## 4 GO_POLY_PURINE_TRACT_BINDING 19 Up
## 5 GO_SINGLE_STRANDED_RNA_BINDING 68 Up
## PValue FDR
## 1 5.726050e-13 5.159171e-10
## 2 9.743160e-12 4.389294e-09
## 3 1.541395e-10 4.629322e-08
## 4 7.946634e-07 1.789979e-04
## 5 1.190113e-05 2.144584e-03
## Cluster: beta Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 6870
## category NGenes Direction
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up
## 2 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_ 18 Up
## 3 GO_POLY_A_BINDING 13 Up
## 4 GO_TRANSLATION_ELONGATION_FACTOR_ACTIVITY 19 Up
## 5 GO_POLY_PURINE_TRACT_BINDING 19 Up
## 6 GO_HISTONE_LYSINE_N_METHYLTRANSFERASE_ACTIVITY 45 Up
## 7 GO_SINGLE_STRANDED_RNA_BINDING 68 Up
## PValue FDR
## 1 4.161799e-24 3.749781e-21
## 2 1.923887e-14 8.667110e-12
## 3 5.219789e-07 1.567677e-04
## 4 3.728563e-05 8.398587e-03
## 5 8.560856e-05 1.212894e-02
## 6 9.094395e-05 1.212894e-02
## 7 9.423149e-05 1.212894e-02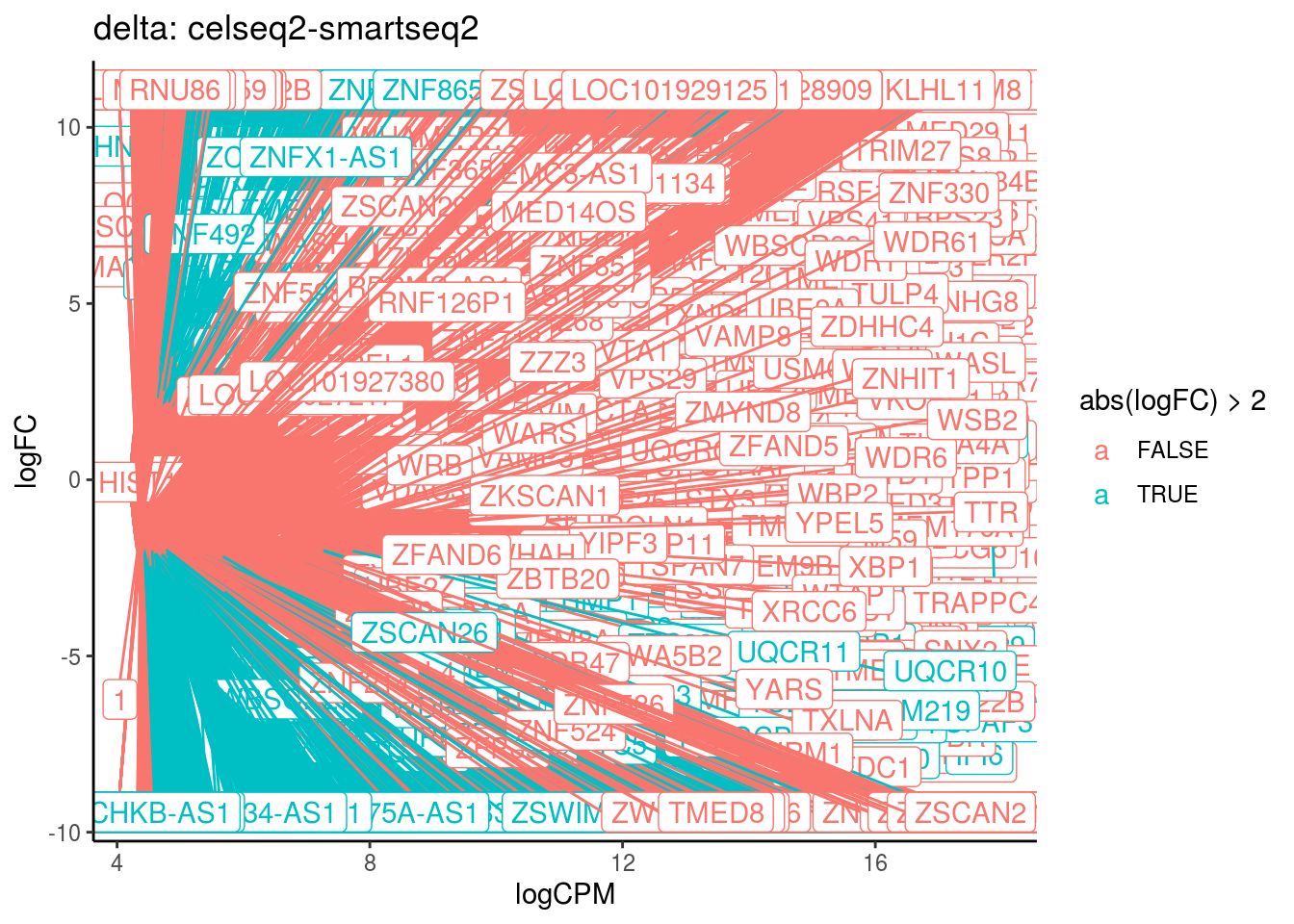
## Cluster: delta Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 7596
## category
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 2 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_
## 3 GO_POLY_A_BINDING
## 4 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 5 GO_HYDROGEN_EXPORTING_ATPASE_ACTIVITY
## 6 GO_GLUTATHIONE_PEROXIDASE_ACTIVITY
## 7 GO_POLY_PURINE_TRACT_BINDING
## 8 GO_HISTONE_LYSINE_N_METHYLTRANSFERASE_ACTIVITY
## NGenes Direction PValue FDR
## 1 204 Up 1.810597e-14 1.631348e-11
## 2 18 Up 5.135129e-14 2.313376e-11
## 3 13 Up 5.652473e-07 1.697626e-04
## 4 52 Up 1.082599e-06 2.438555e-04
## 5 27 Up 1.869598e-06 3.369016e-04
## 6 17 Up 1.101936e-04 1.654741e-02
## 7 19 Up 1.664670e-04 2.142668e-02
## 8 45 Up 2.182002e-04 2.457480e-02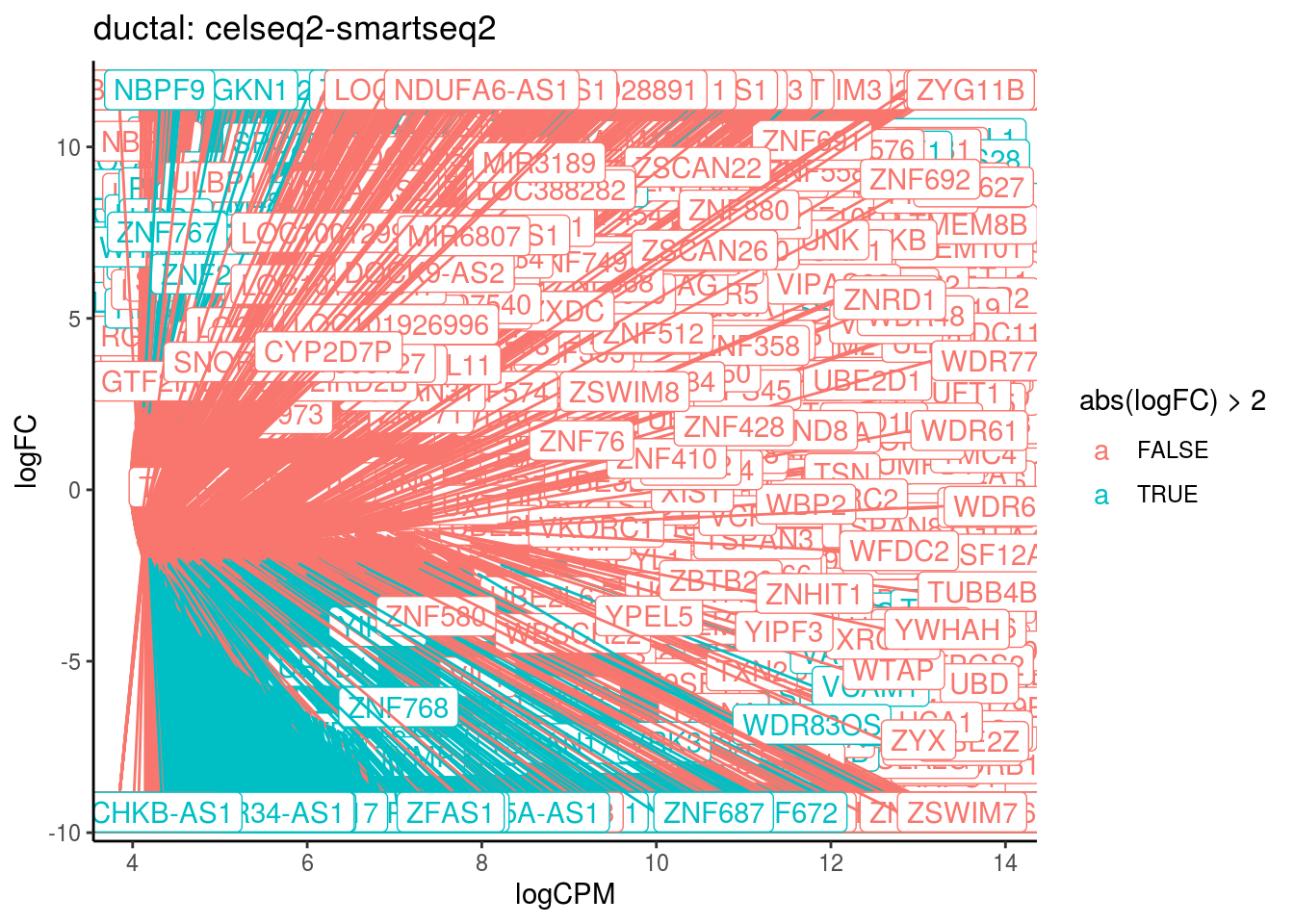
## Cluster: ductal Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 7017
## category NGenes Direction
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up
## 2 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_ 18 Up
## 3 GO_POLY_A_BINDING 13 Up
## 4 GO_POLY_PURINE_TRACT_BINDING 19 Up
## 5 GO_SINGLE_STRANDED_RNA_BINDING 68 Up
## 6 GO_HISTONE_LYSINE_N_METHYLTRANSFERASE_ACTIVITY 45 Up
## 7 GO_LYSINE_N_METHYLTRANSFERASE_ACTIVITY 55 Up
## 8 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY 58 Up
## PValue FDR
## 1 1.688937e-20 1.521732e-17
## 2 8.435694e-15 3.800280e-12
## 3 3.448317e-11 1.035645e-08
## 4 2.976008e-07 6.703457e-05
## 5 2.006862e-06 3.616365e-04
## 6 1.332967e-05 2.001672e-03
## 7 1.214480e-04 1.563209e-02
## 8 2.425620e-04 2.731855e-02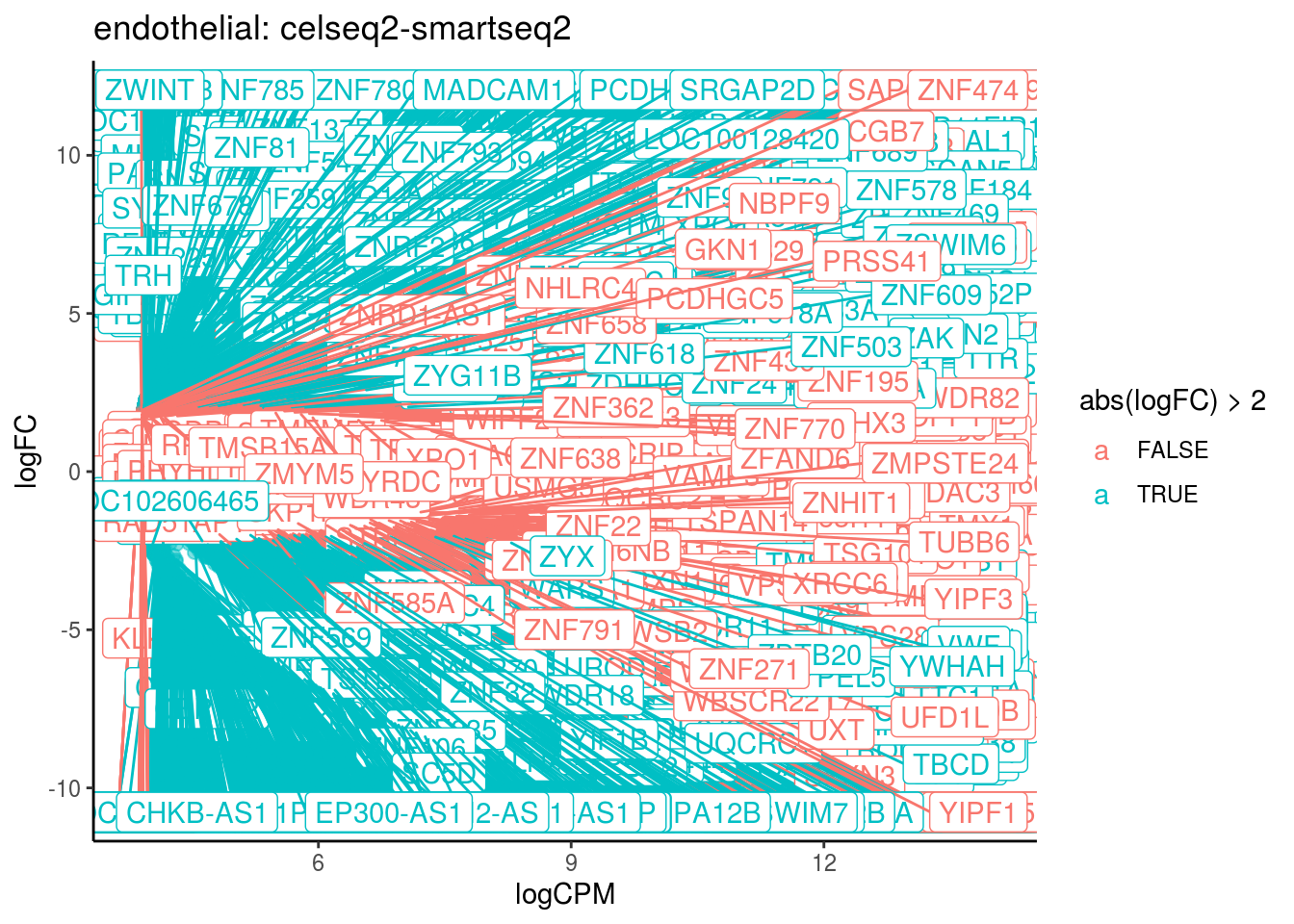
## Cluster: endothelial Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 5346
## category
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 2 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_
## 3 GO_SINGLE_STRANDED_RNA_BINDING
## 4 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 5 GO_POLY_A_BINDING
## 6 GO_THREONINE_TYPE_PEPTIDASE_ACTIVITY
## 7 GO_DNA_POLYMERASE_BINDING
## 8 GO_HISTONE_LYSINE_N_METHYLTRANSFERASE_ACTIVITY
## NGenes Direction PValue FDR
## 1 204 Up 3.093825e-14 2.787536e-11
## 2 18 Up 1.985799e-12 8.946027e-10
## 3 68 Up 4.103683e-06 1.232473e-03
## 4 52 Up 8.958421e-06 2.017884e-03
## 5 13 Up 1.193483e-05 2.150656e-03
## 6 21 Up 1.275931e-04 1.916023e-02
## 7 12 Up 1.869623e-04 2.406472e-02
## 8 45 Up 3.352597e-04 3.775863e-02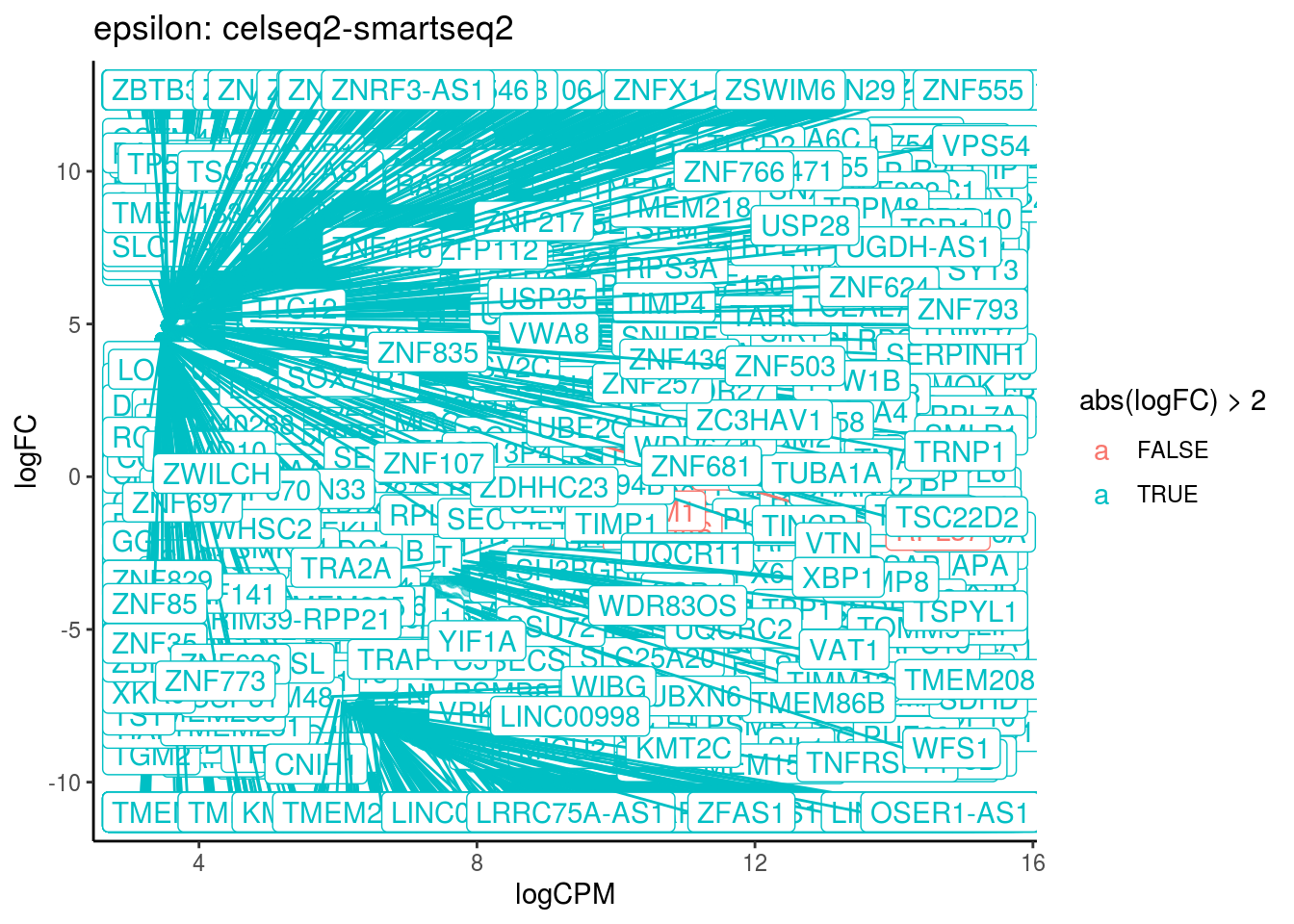
## Cluster: epsilon Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 1431
## category
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 2 GO_TRANSLATION_ELONGATION_FACTOR_ACTIVITY
## 3 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 4 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_
## 5 GO_DIPEPTIDYL_PEPTIDASE_ACTIVITY
## NGenes Direction PValue FDR
## 1 204 Up 1.414038e-12 1.274049e-09
## 2 19 Up 2.893433e-06 1.303491e-03
## 3 52 Up 2.148590e-05 6.452931e-03
## 4 18 Up 3.925370e-05 8.841895e-03
## 5 13 Up 5.661013e-05 1.020114e-02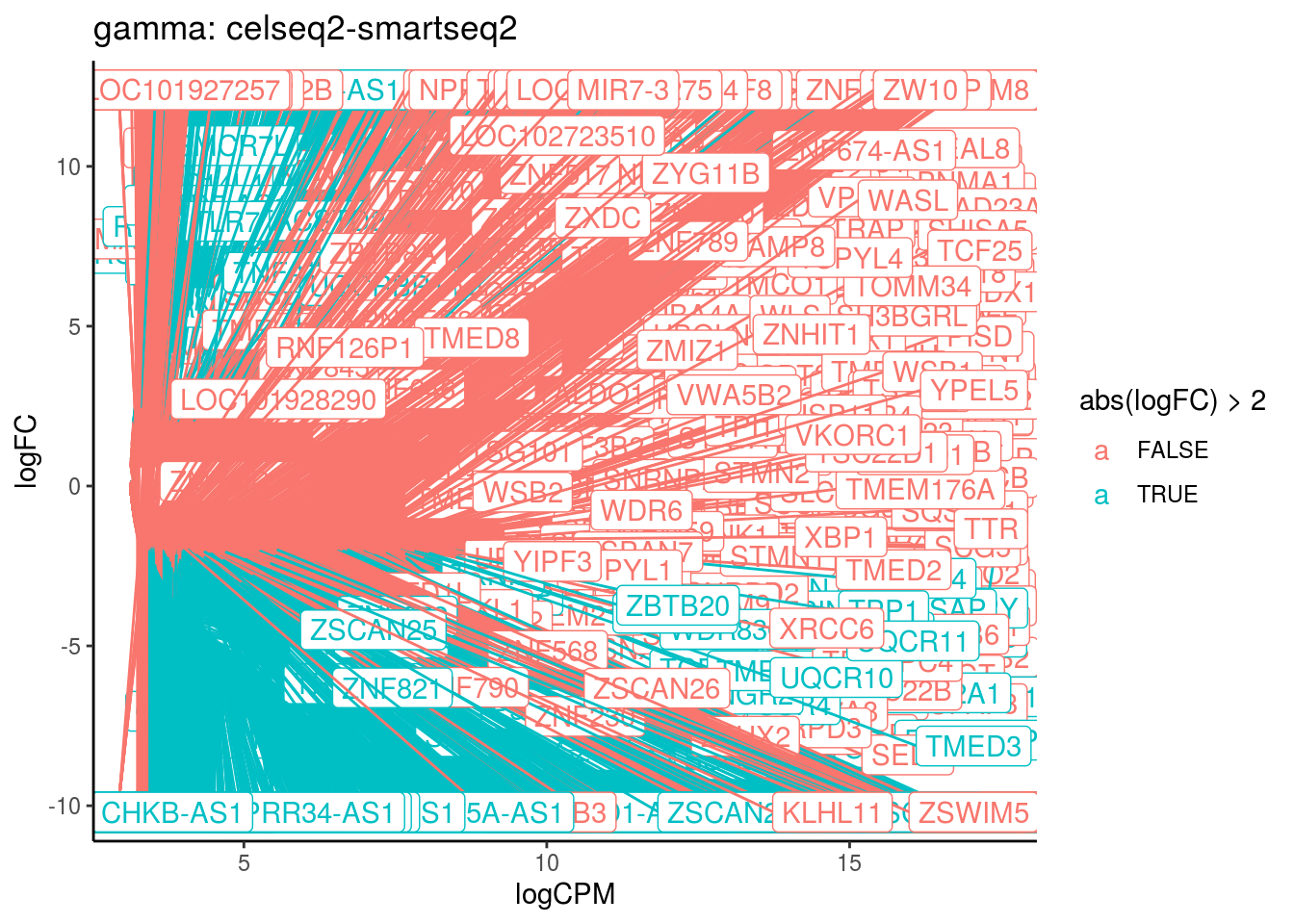
## Cluster: gamma Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 8476
## category NGenes Direction
## 1 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_ 18 Up
## 2 GO_POLY_A_BINDING 13 Up
## 3 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up
## PValue FDR
## 1 4.585539e-11 4.131571e-08
## 2 6.036089e-06 2.719258e-03
## 3 2.835032e-05 8.514545e-03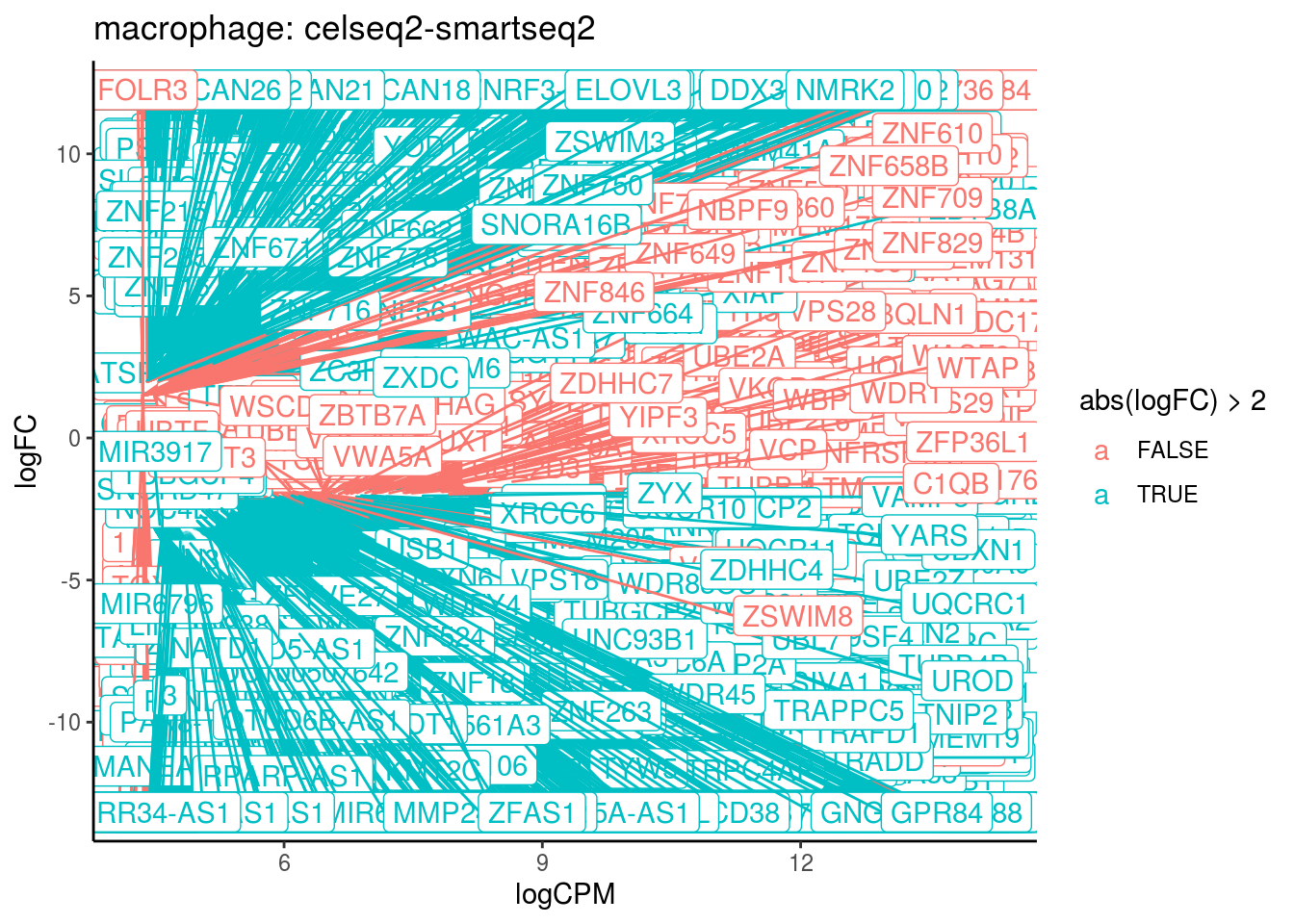
## Cluster: macrophage Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 4687
## category
## 1 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_
## 2 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME
## 3 GO_PEPTIDE_ANTIGEN_BINDING
## 4 GO_HYDROGEN_EXPORTING_ATPASE_ACTIVITY
## 5 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H_QUINONE_OR_SIMILAR_COMPOUND_AS_ACCEPTOR
## 6 GO_THREONINE_TYPE_PEPTIDASE_ACTIVITY
## 7 GO_MHC_CLASS_II_RECEPTOR_ACTIVITY
## 8 GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_NAD_P_H
## NGenes Direction PValue FDR
## 1 18 Up 6.066051e-12 5.465512e-09
## 2 204 Up 3.663023e-11 1.650192e-08
## 3 27 Up 1.788458e-09 5.371337e-07
## 4 27 Up 6.800107e-09 1.531724e-06
## 5 52 Up 2.818619e-08 5.079151e-06
## 6 21 Up 4.414449e-07 6.412972e-05
## 7 10 Up 4.982331e-07 6.412972e-05
## 8 93 Up 5.740387e-07 6.465110e-05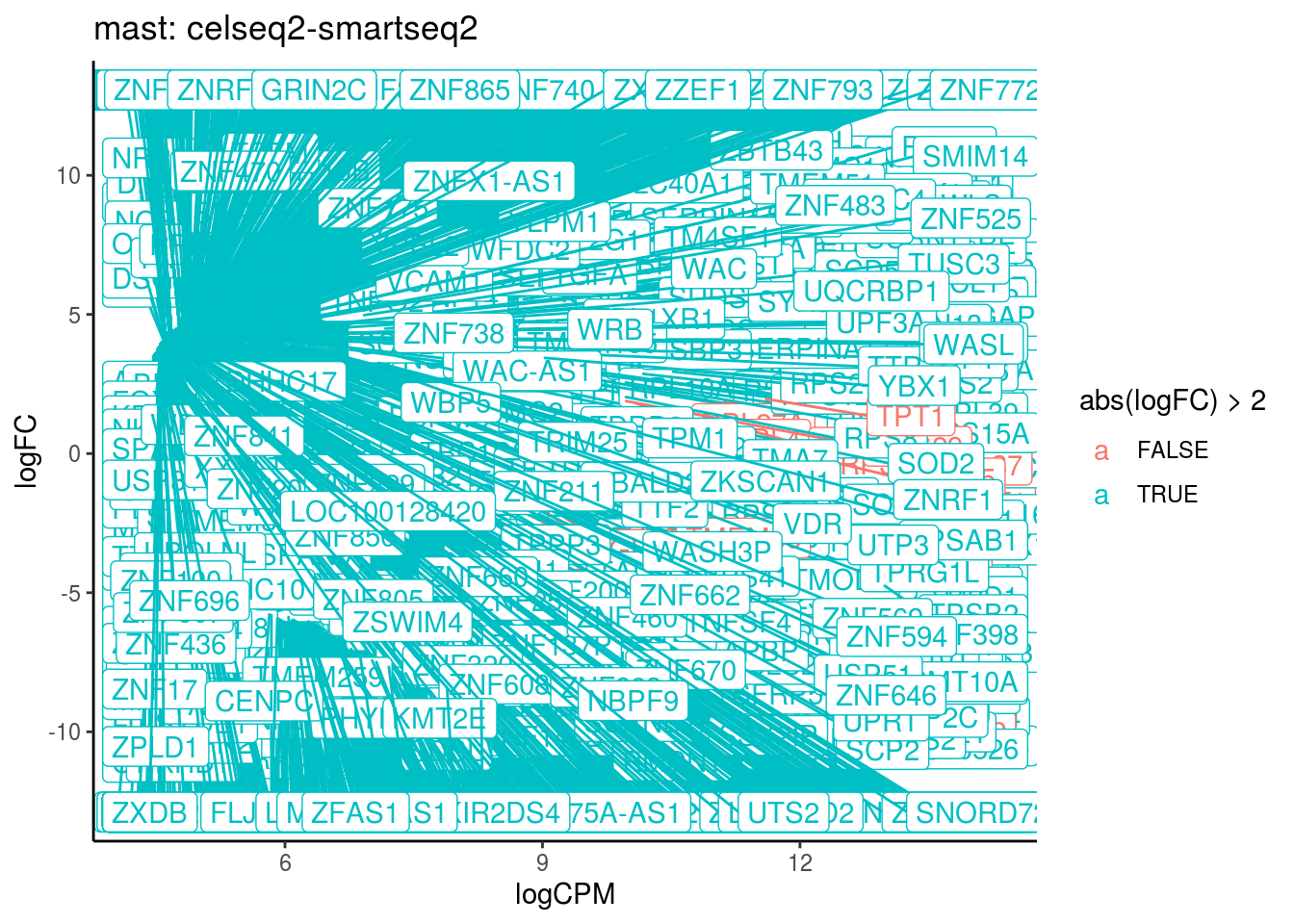
## Cluster: mast Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 3443
## category NGenes Direction PValue
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up 4.161615e-16
## 2 GO_GLUTATHIONE_PEROXIDASE_ACTIVITY 17 Up 8.109805e-06
## FDR
## 1 3.749615e-13
## 2 3.653467e-03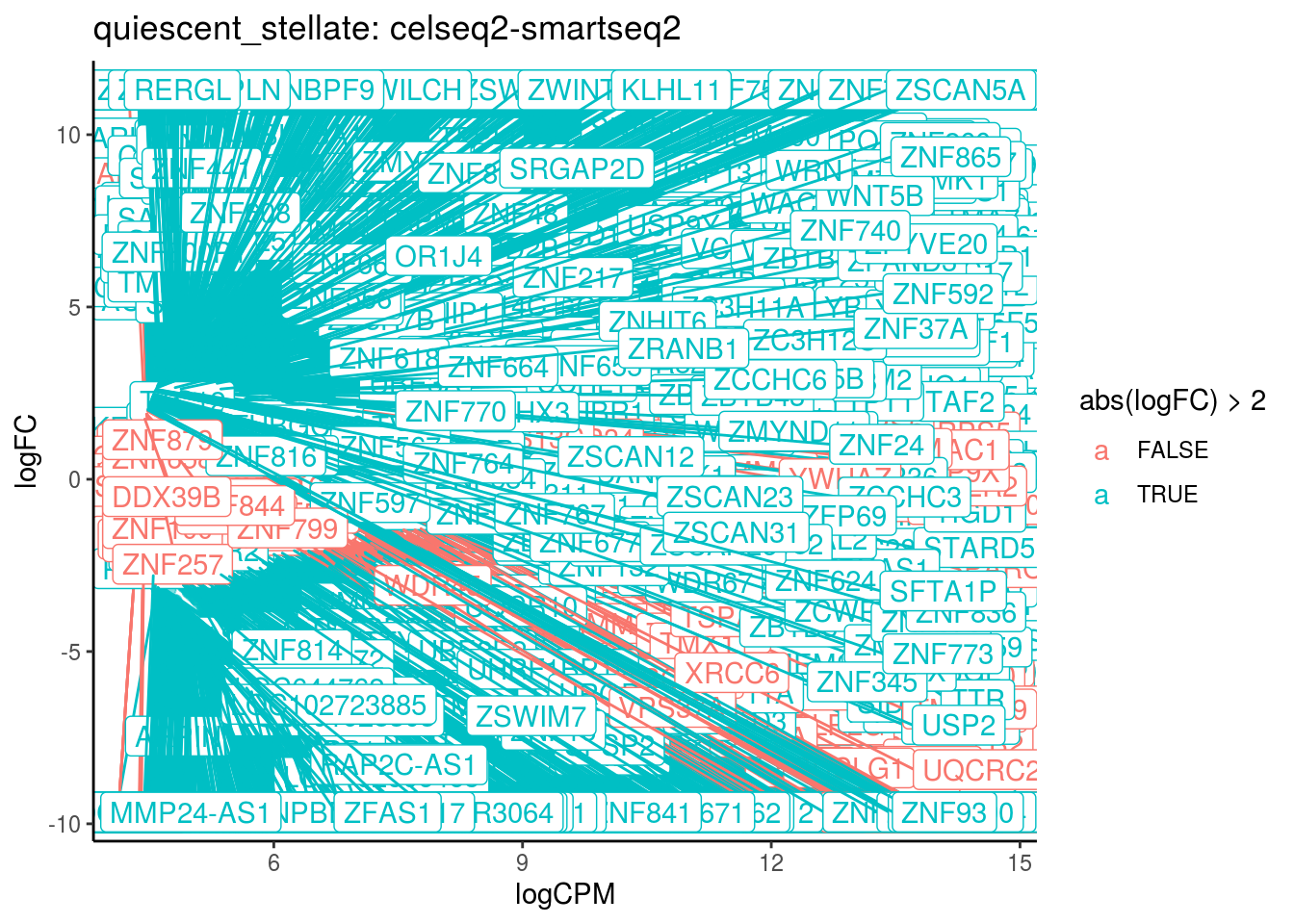
## Cluster: quiescent_stellate Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 5283
## category NGenes Direction
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up
## 2 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_ 18 Up
## 3 GO_POLY_A_BINDING 13 Up
## 4 GO_POLY_PURINE_TRACT_BINDING 19 Up
## 5 GO_HISTONE_LYSINE_N_METHYLTRANSFERASE_ACTIVITY 45 Up
## 6 GO_TRANSFORMING_GROWTH_FACTOR_BETA_BINDING 16 Up
## 7 GO_SINGLE_STRANDED_RNA_BINDING 68 Up
## 8 GO_GLUTATHIONE_PEROXIDASE_ACTIVITY 17 Up
## PValue FDR
## 1 4.177346e-23 3.763789e-20
## 2 8.043816e-18 3.623739e-15
## 3 2.471650e-11 7.423189e-09
## 4 1.114716e-10 2.510897e-08
## 5 2.655449e-09 4.785119e-07
## 6 8.437335e-09 1.267006e-06
## 7 1.626842e-08 2.093978e-06
## 8 4.206412e-08 4.737472e-06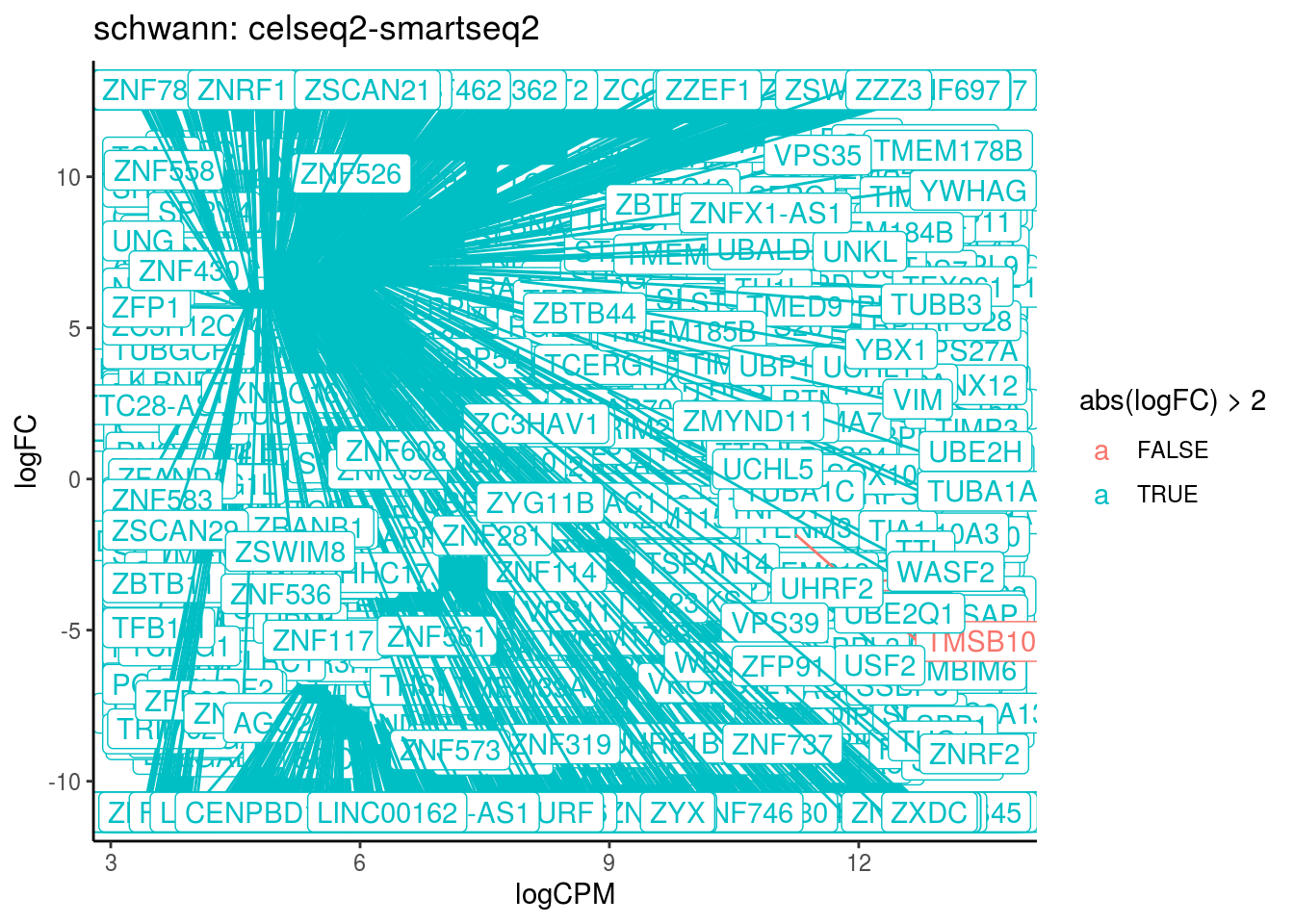
## Cluster: schwann Contrast: celseq2-smartseq2 Num genes: 24509 Num DE: 2762
## category NGenes Direction
## 1 GO_STRUCTURAL_CONSTITUENT_OF_RIBOSOME 204 Up
## 2 GO_POLY_A_BINDING 13 Up
## 3 GO_PROTEOGLYCAN_BINDING 29 Up
## 4 GO_POLY_PURINE_TRACT_BINDING 19 Up
## 5 GO_POLY_A_RNA_BINDING 1149 Up
## 6 GO_HISTONE_METHYLTRANSFERASE_ACTIVITY_H3_K4_SPECIFIC_ 18 Up
## 7 GO_SINGLE_STRANDED_RNA_BINDING 68 Up
## 8 GO_MRNA_BINDING 152 Up
## PValue FDR
## 1 1.711473e-18 1.542037e-15
## 2 1.306536e-11 5.885943e-09
## 3 4.842051e-11 1.454229e-08
## 4 1.645907e-10 3.707406e-08
## 5 1.183986e-07 2.133543e-05
## 6 1.641627e-07 2.143182e-05
## 7 1.665069e-07 2.143182e-05
## 8 4.715516e-07 5.310850e-05Summarize differential expression analysis
# DE genes (per cluster and mean)
#n_de <- lapply(res, function(y) vapply(y, function(x) sum(x$adj.P.Val < 0.05), numeric(1)))
n_de <- lapply(res, function(y) vapply(y, function(x) sum(x$PValue < 0.05), numeric(1)))
n_de_cl <- lapply(res, function(y) vapply(y, function(x) nrow(x), numeric(1)))
mean_n_de <- lapply(n_de, function(x) mean(x))
mean_mean_n_de <- mean(unlist(mean_n_de))/n_genes
min_mean_n_de <- min(unlist(mean_n_de))/n_genes
max_mean_n_de <- max(unlist(mean_n_de))/n_genes
# Genes with lfc > 1
n_genes_lfc1 <- lapply(res, function(y) vapply(y, function(x) sum(abs(x$logFC) > 1), numeric(1)))
mean_n_genes_lfc1 <- mean(unlist(n_genes_lfc1))/n_genes
min_n_genes_lfc1 <- min(unlist(n_genes_lfc1))/n_genes
max_n_genes_lfc1 <- max(unlist(n_genes_lfc1))/n_genes
# DE genes overlap between celltypes (celltype specific de genes)
# Genes are "overlapping" if they are present in all clusters with at least 10% of all cells
de_overlap <- lapply(result, function(x){
result2 <- x[table(colData(sce)[, celltype]) > ncol(sce) * 0.1]
de_overlap <- length(Reduce(intersect, result2))
de_overlap
})
mean_de_overlap <- mean(unlist(de_overlap))/n_genes
min_de_overlap <- min(unlist(de_overlap))/n_genes
max_de_overlap <- max(unlist(de_overlap))/n_genes
#Genes unique to single celltypes
unique_genes_matrix <- NULL
unique_genes <- NULL
cb <- length(names(result[[1]]))
unique_genes <- lapply(result,function(x){
for( i in 1:cb ){
unique_genes[i] <-as.numeric(length(setdiff(unlist(x[i]),unlist(x[-i]))))
}
unique_genes_matrix <- cbind(unique_genes_matrix, unique_genes)
unique_genes_matrix
})
unique_genes <- Reduce('cbind', unique_genes)
colnames(unique_genes) <- names(result)
rownames(unique_genes) <- names(result[[1]])
# Relative cluster specificity (unique/overlapping)
rel_spec1 <- NULL
for( i in 1:dim(unique_genes)[2] ){
rel_spec <- unique_genes[,i]/de_overlap[[i]]
rel_spec1 <- cbind(rel_spec1,rel_spec)
}
mean_rel_spec <- mean(rel_spec1)
min_rel_spec <- min(rel_spec1)
max_rel_spec <- max(rel_spec1)Celltype specific DE distributions
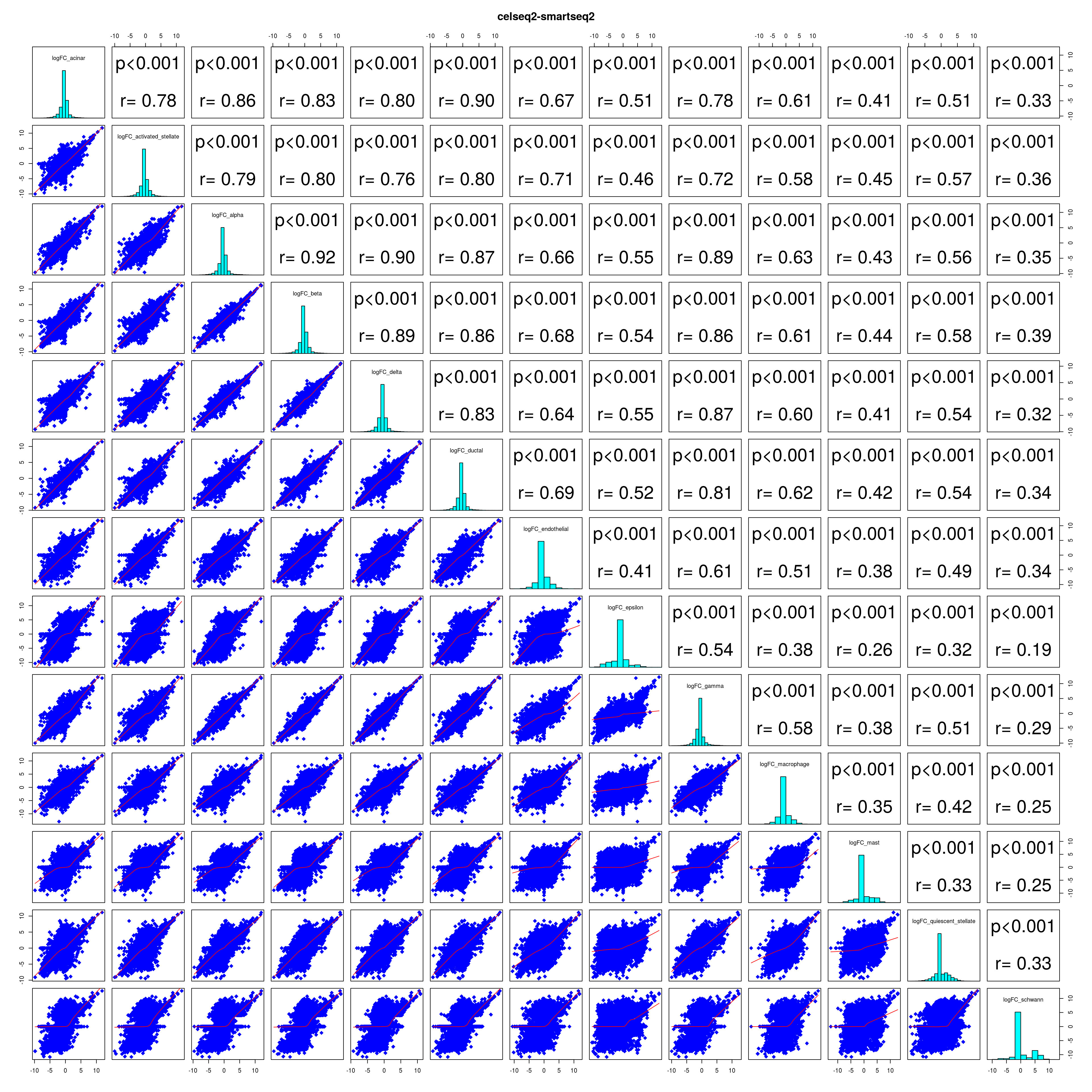
How similar is the batch effect between celltypes. Do we have similar logFC distributions or different?
combine_folds <- function(cont_var){
#extract the contrast of interest and change log2fold colums names to be unique
B <- res[[cont_var]]
new_name <- function(p){
colnames(B[[p]])[3] <- paste0("logFC_", p)
return(B[[p]][,c(1,3)])
}
B_new_names <- lapply(names(B),new_name)
names(B_new_names) <- names(B)
#combine log2fold colums
Folds <- Reduce(function(...){inner_join(..., by="gene")}, B_new_names)
}
all_folds <- lapply(cs, combine_folds)
#define pannels for pairs() function
panel.cor <- function(x, y, digits = 2, cex.cor){
usr <- par("usr"); on.exit(par(usr))
par(usr = c(0, 1, 0, 1))
r <- abs(cor(x, y))
txt <- format(c(r, 0.123456789), digits=digits)[1]
test <- cor.test(x,y)
Signif <- ifelse(round(test$p.value, 3) < 0.001,
"p<0.001",
paste("p=",round(test$p.value,3)))
text(0.5, 0.25, paste("r=",txt), cex = 3)
text(.5, .75, Signif, cex = 3)
}
panel.smooth <- function (x, y, col = "blue", bg = NA, pch = 18, cex = 1.5,
col.smooth = "red", span = 2/3, iter = 3, ...){
points(x, y, pch = pch, col = col, bg = bg, cex = cex)
ok <- is.finite(x) & is.finite(y)
if( any(ok) )
lines(stats::lowess(x[ok], y[ok], f = span, iter = iter),
col = col.smooth, ...)
}
panel.hist <- function(x, ...){
usr <- par("usr"); on.exit(par(usr))
par(usr = c(usr[1:2], 0, 1.5) )
h <- hist(x, plot = FALSE)
breaks <- h$breaks
nB <- length(breaks)
y <- h$counts
y <- y/max(y)
rect(breaks[-nB], 0, breaks[-1], y, col="cyan", ...)
}
#plot correlations
lapply(names(all_folds), function(x) pairs(all_folds[[x]][,-1],
lower.panel = panel.smooth,
upper.panel = panel.cor,
diag.panel = panel.hist, main = x))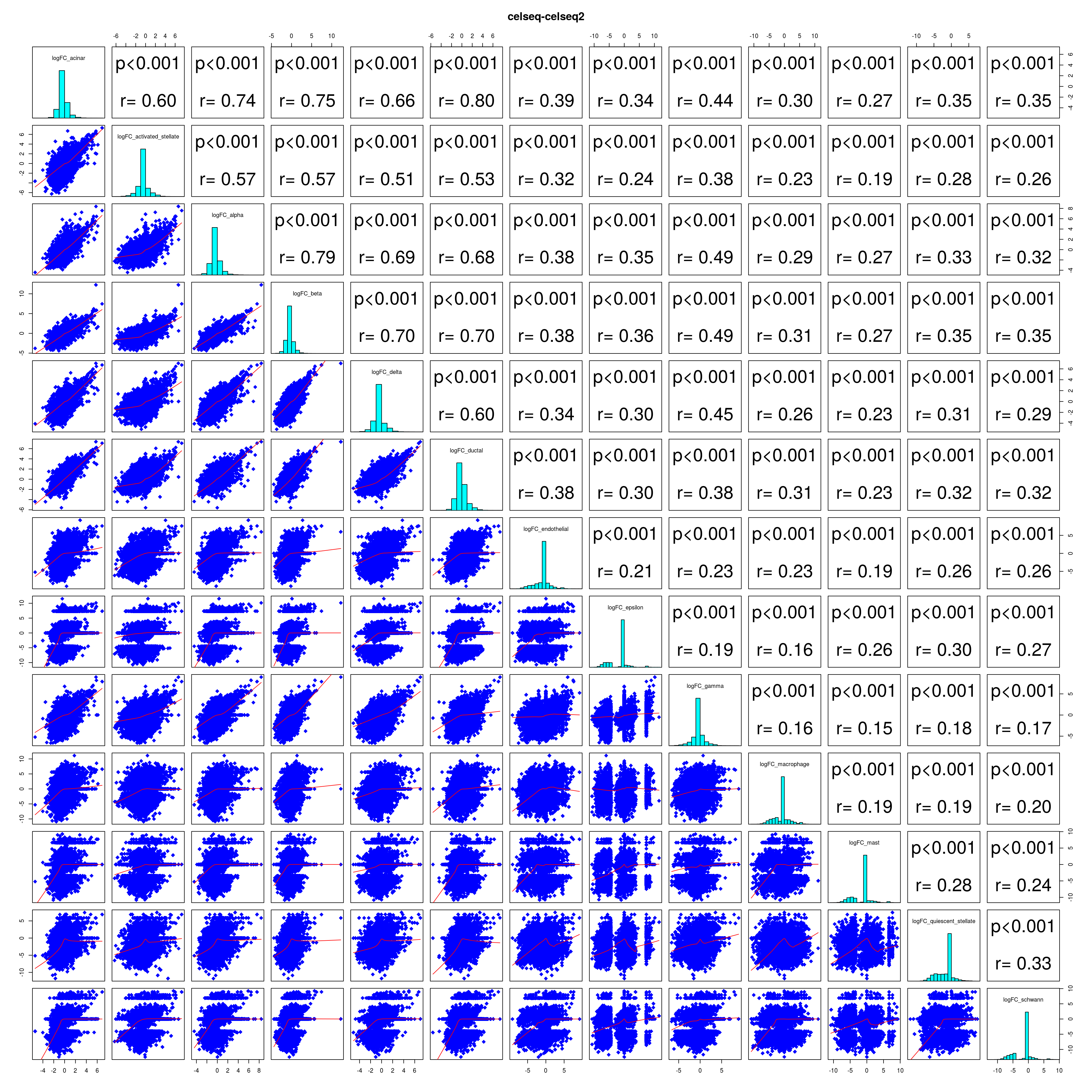

## [[1]]
## NULL
##
## [[2]]
## NULL
##
## [[3]]
## NULL#extract correlation coefficients
# correlation coefficients from celltype specific gege logFC
lfc_cor_list <-lapply(names(all_folds), function(com){
exclude <- which(table(colData(sce)[,celltype]) < 100)
r <- cor(all_folds[[com]][, -c(1, (exclude + 1))])
mean_r <- (sum(r) - ncol(r))/ (ncol(r)^2 - ncol(r))
})
mean_lfc_cor <- mean(unlist(lfc_cor_list))Batch categorization
How does the batch effect manifest? Can we describe it by “simple” mean shifts of expression levels for some genes for all the cells in a given celltype and batch? Can we “remove” the batch effcet using a linear model with batch, batch and celltype or batch and celltype interacting?
#Visualize different models
vis_type <- function(dim_red){
g <- visGroup(sce, batch, dim_red = dim_red) +
ggtitle("unadjusted")
g1 <- visGroup(sce, batch, dim_red = paste0(dim_red, "_Xadj1")) +
ggtitle("constant batch effect")
g2 <- visGroup(sce, batch, dim_red = paste0(dim_red, "_Xadj2")) +
ggtitle("constant batch effect, different ct composition")
g3 <- visGroup(sce, batch, dim_red = paste0(dim_red, "_Xadj3")) +
ggtitle("celltype and batch effect interact")
do.call("grid.arrange", c(list(g, g1, g2, g3), ncol = 2))
}PCA
vis_type("PCA")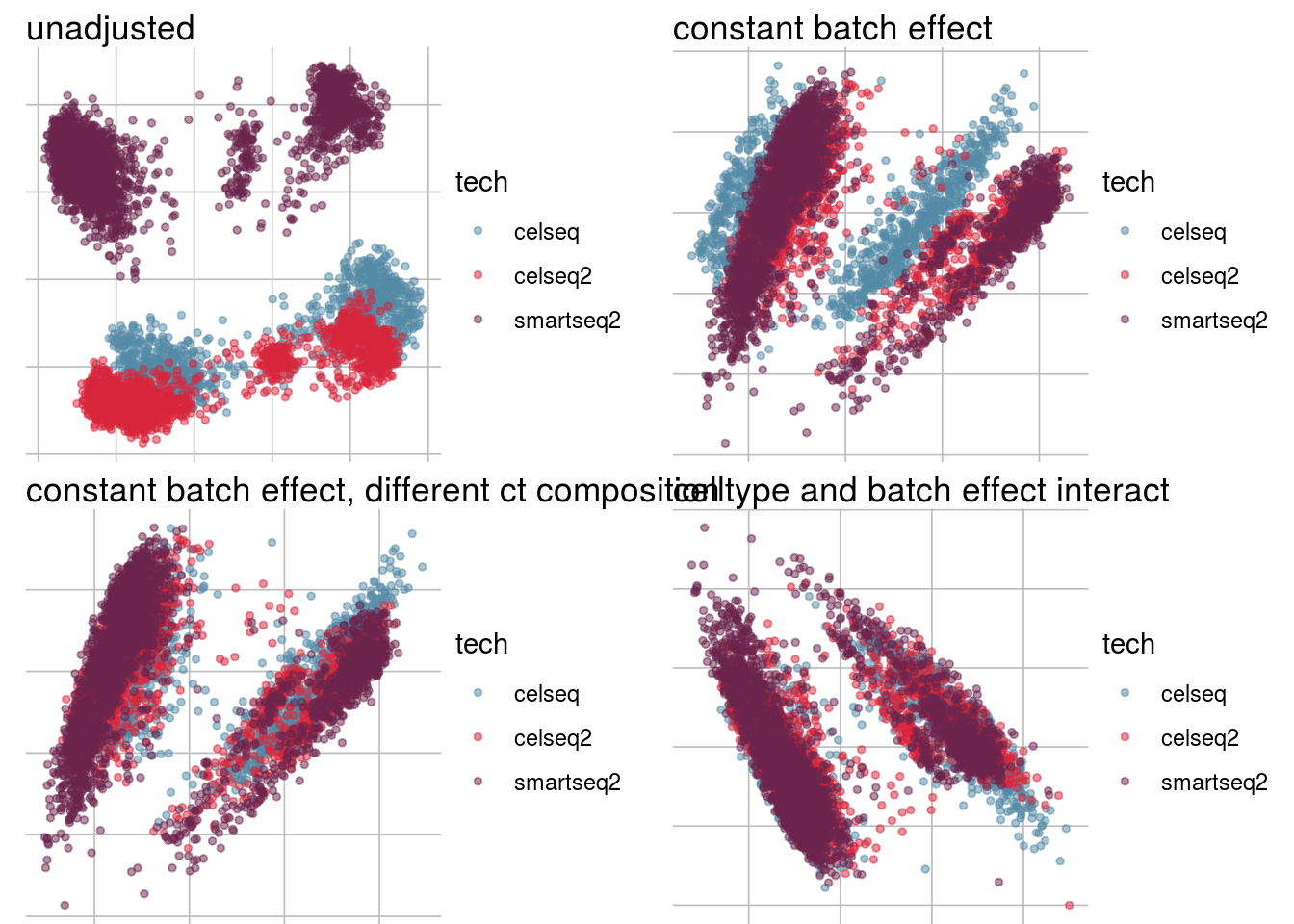
UMAP
vis_type("UMAP")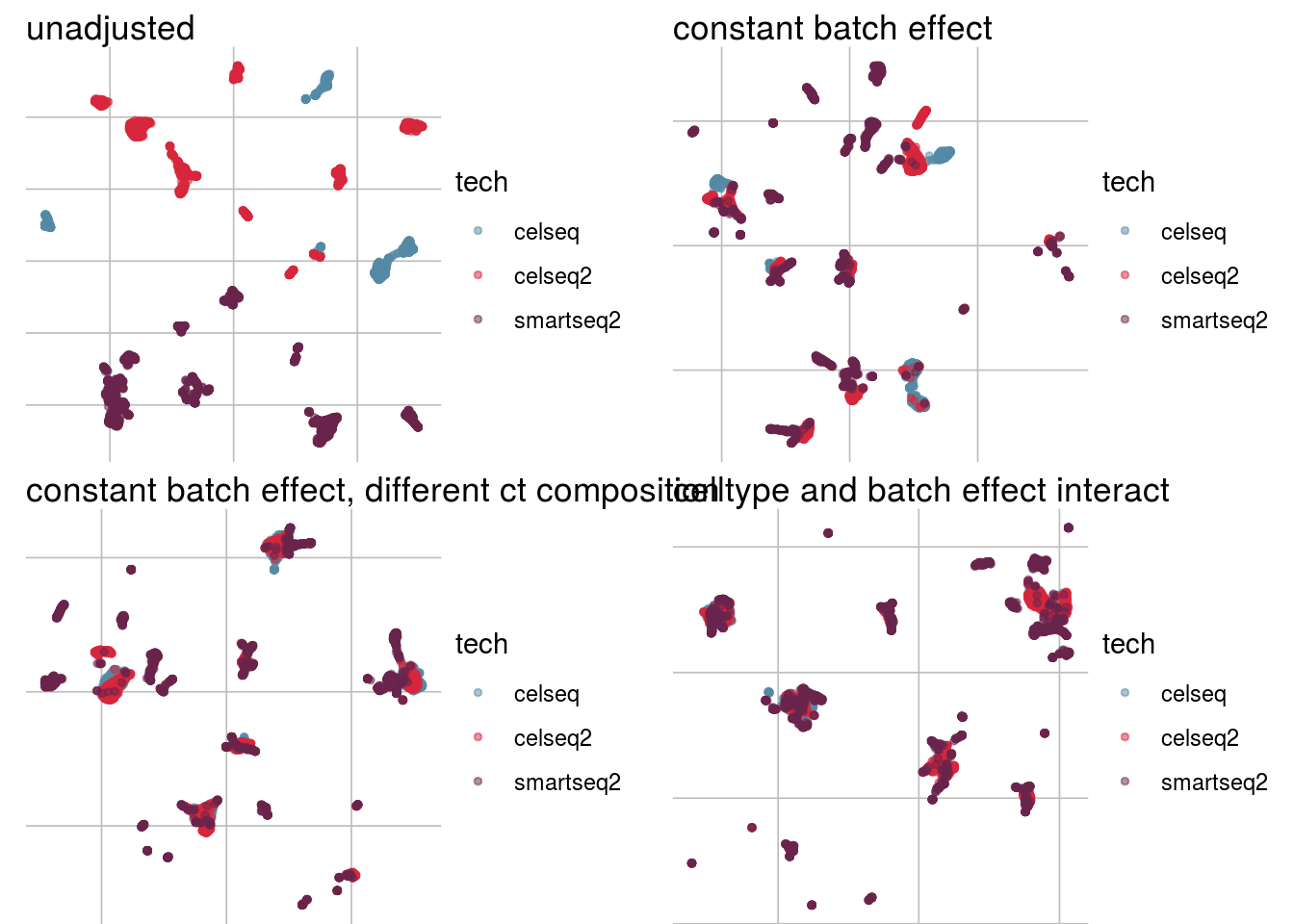
Cellspecific Mixing Score
# #Cellspecific Mixing score (Batch effect strength after "removal")
visHist(sce, metric = c("cms", "cms.Xadj1", "cms.Xadj2", "cms.Xadj3"), prefix = FALSE)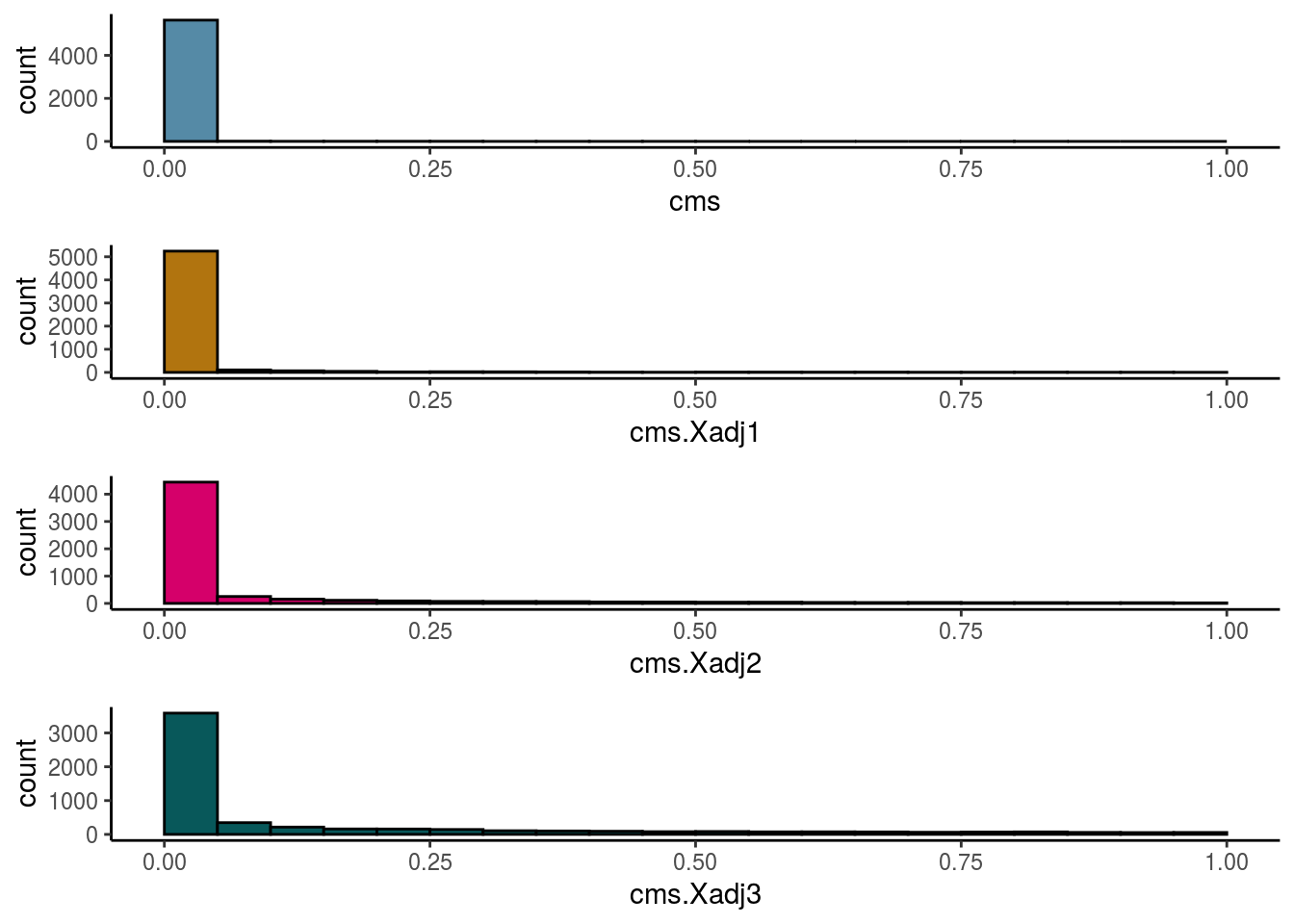
visIntegration(sce, metric = "cms", metric_name = "cms")## Picking joint bandwidth of 0.00849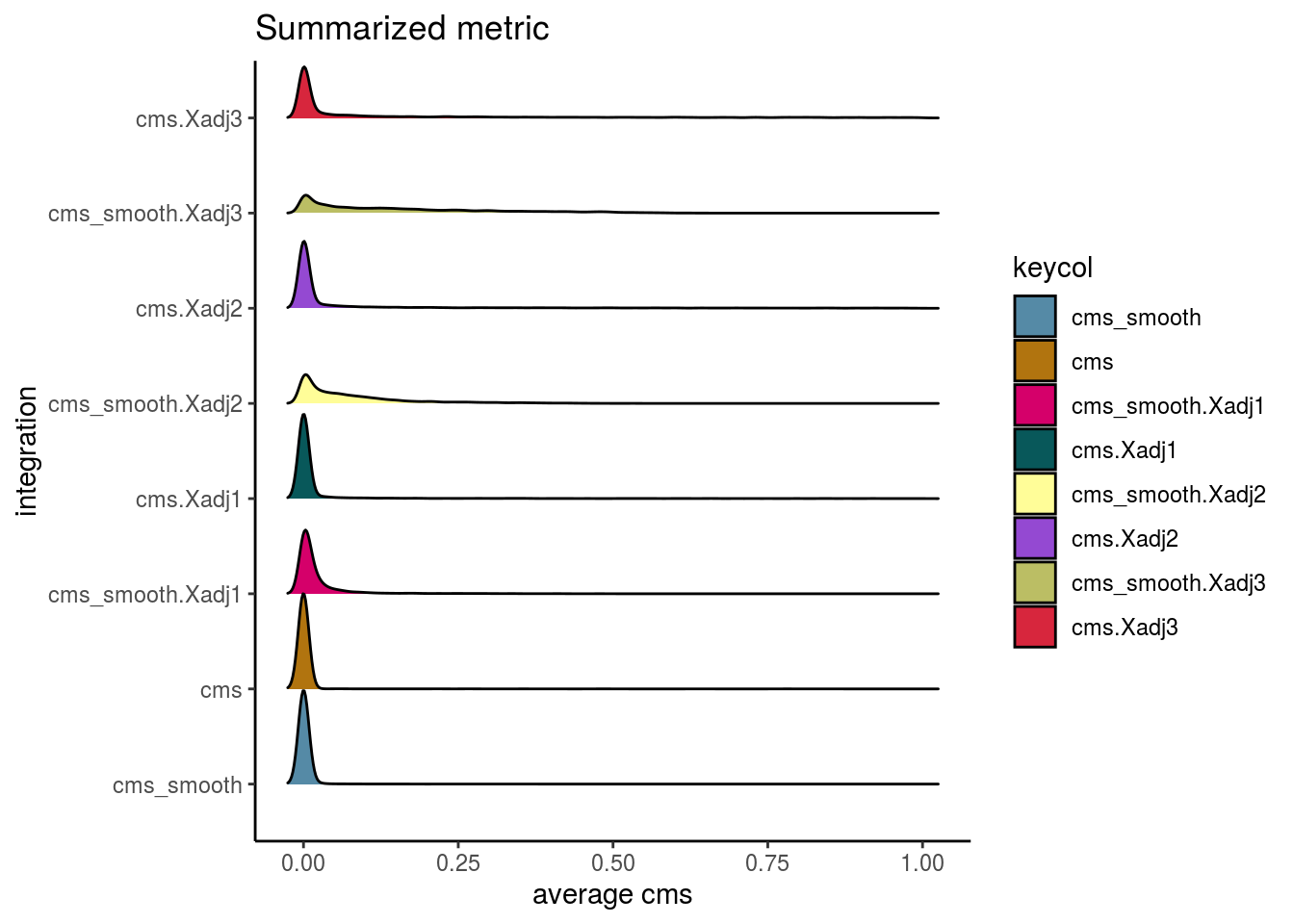
Simulation parameter
Extract parameter to use as input into simualation
#percentage of batch affected genes
cond <- gsub("-.*", "", names(n_de))
cond <- c(cond, unique(gsub(".*-", "", names(n_de))))
cond <- unique(cond)
de_be_tab <- n_de %>% bind_cols()
de_cl_tab <- n_de_cl %>% bind_cols()
de_be <- cond %>% map(function(x){
de_tab <- de_be_tab[, grep(x, colnames(de_be_tab))]
de_be <- rowMeans(de_tab)
}) %>% bind_cols() %>% set_colnames(cond)
n_cl <- cond %>% map(function(x){
cl_tab <- de_cl_tab[, grep(x, colnames(de_cl_tab))]
de_cl <- rowMeans(cl_tab)
}) %>% bind_cols() %>% set_colnames(cond)
p_be <- de_be/n_cl
mean_p_be <- mean(colMeans(p_be))
min_p_be <- min(colMins(as.matrix(p_be)))
max_p_be <- max(colMaxs(as.matrix(p_be)))
sd_p_be <- mean(colSds(as.matrix(p_be)))
if(is.na(sd_p_be)){ sd_p_be <- 0 }
#### Percentage of celltype specific genes "p_ct"
n_de_unique <- lapply(result,function(x){
de_genes <- unlist(x) %>% unique() %>% length()
de_genes <- de_genes/length(x)
}) %>% bind_cols()
rel_spec2 <- NULL
for(i in 1:length(de_overlap)){
rel_spec <- de_overlap[[i]]/mean(n_de[[i]][table(colData(sce)[, celltype]) > dim(expr)[2] * 0.1])
rel_spec2 <- cbind(rel_spec2, rel_spec)
}
mean_p_ct <- 1 - mean(rel_spec2)
max_p_ct <- 1 - min(rel_spec2)
min_p_ct <- 1 - max(rel_spec2)
sd_p_ct <- sd(rel_spec2)
if(is.na(sd_p_ct)){ sd_p_ct <- 0 }
# Logfold change
#logFoldchange batch effect distribution
mean_lfc_cl <- lapply(res, function(y) vapply(y, function(x){
#de_genes <- which(x$adj.P.Val < 0.05)
de_genes <- which(x$adj.PValue < 0.05)
mean_de <- mean(abs(x[, "logFC"]))}
, numeric(1))) %>% bind_cols()
mean_lfc_be <- mean(colMeans(mean_lfc_cl, na.rm = TRUE))
min_lfc_be <- min(colMins(as.matrix(mean_lfc_cl), na.rm = TRUE))
max_lfc_be <- max(colMaxs(as.matrix(mean_lfc_cl), na.rm = TRUE))Summarize batch effect
- Batch size
- Celltype specificity
- “Batch genes”
- batch type
#Size? How much of the variance can be attributed to the batch effect?
size <- data.frame("batch_genes_1per" = n_batch_gene, # 1.variance partition
"batch_genes_10per" = n_batch_gene10,
"celltype_gene_1per" = n_celltype_gene,
"relative_batch_celltype" = n_rel,
"mean_var_batch" = m_batch,
"mean_var_celltype" = m_celltype,
"rel_mean_ct_batch" = m_rel,
"mean_cms" = mean_cms, #2.cms
"n_cells_cms_0.01" = n_cms_0.01,
"mean_mean_n_de_genes" = mean_mean_n_de, #3.de genes
"max_mean_n_de_genes" = max_mean_n_de,
"min_mean_n_de_genes" = min_mean_n_de,
"mean_n_genes_lfc1" = mean_n_genes_lfc1,
"min_n_genes_lfc1" = min_n_genes_lfc1,
"max_n_genes_lfc1" = max_n_genes_lfc1,
"n_cells_total" = ncol(sce), #4.general
"n_genes_total" = nrow(sce))
#Celltype-specificity? How celltype/cluster specific are batch effects?
# Differences in size, distribution or abundance? Do we find correlations between lfcs,
# overlap in de genes, pathways? Interaction between ct and be?
celltype <- data.frame('mean_rel_abund_diff' = mean_rel_abund_diff, #1.abundance
'min_rel_abund_diff' = min_rel_abund_diff,
'max_rel_abund_diff' = max_rel_abund_diff,
"celltype_var_cms" = var_cms, #2.size/strength
"mean_de_overlap" = mean_de_overlap,
"min_de_overlap" = min_de_overlap,
"max_de_overlap" = max_de_overlap,
"mean_rel_cluster_spec"= mean_rel_spec,
"min_rel_cluster_spec"= min_rel_spec,
"max_rel_cluster_spec"= max_rel_spec,
"mean_lfc_cor" = mean_lfc_cor )
sim <- data.frame("mean_p_be" = mean_p_be,
"max_p_be" = max_p_be,
"min_p_be" = min_p_be,
"sd_p_be" = sd_p_be,
"mean_lfc_be" = mean_lfc_be,
"min_lfc_be" = min_lfc_be,
"max_lfc_be" = max_lfc_be,
"mean_p_ct"= mean_p_ct,
"min_p_ct"= min_p_ct,
"max_p_ct"= max_p_ct,
"sd_p_ct" = sd_p_ct)
summary <- cbind(size, celltype, sim) %>% set_rownames(dataset_name)
### -------------- save summary object ----------------------###
saveRDS(summary, file = outputfile)sessionInfo()## R version 3.6.1 (2019-07-05)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 16.04.6 LTS
##
## Matrix products: default
## BLAS: /home/aluetg/R/lib/R/lib/libRblas.so
## LAPACK: /home/aluetg/R/lib/R/lib/libRlapack.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] grid parallel stats4 stats graphics grDevices utils
## [8] datasets methods base
##
## other attached packages:
## [1] readr_1.3.1 ggrepel_0.8.2
## [3] CAMERA_1.42.0 xcms_3.8.2
## [5] MSnbase_2.12.0 ProtGenerics_1.18.0
## [7] mzR_2.20.0 Rcpp_1.0.3
## [9] cowplot_1.0.0 scran_1.14.6
## [11] gridExtra_2.3 ComplexHeatmap_2.2.0
## [13] stringr_1.4.0 dplyr_0.8.5
## [15] tidyr_1.0.2 here_0.1
## [17] jcolors_0.0.4 purrr_0.3.3
## [19] variancePartition_1.16.1 scales_1.1.0
## [21] foreach_1.4.8 limma_3.42.2
## [23] CellMixS_1.2.4 kSamples_1.2-9
## [25] SuppDists_1.1-9.5 scater_1.14.6
## [27] ggplot2_3.3.0 CellBench_1.2.0
## [29] tibble_2.1.3 magrittr_1.5
## [31] SingleCellExperiment_1.8.0 SummarizedExperiment_1.16.1
## [33] DelayedArray_0.12.2 BiocParallel_1.20.1
## [35] matrixStats_0.55.0 Biobase_2.46.0
## [37] GenomicRanges_1.38.0 GenomeInfoDb_1.22.0
## [39] IRanges_2.20.2 S4Vectors_0.24.3
## [41] BiocGenerics_0.32.0
##
## loaded via a namespace (and not attached):
## [1] tidyselect_1.0.0 lme4_1.1-21 RSQLite_2.2.0
## [4] htmlwidgets_1.5.1 munsell_0.5.0 codetools_0.2-16
## [7] preprocessCore_1.48.0 statmod_1.4.34 withr_2.1.2
## [10] colorspace_1.4-1 knitr_1.28 rstudioapi_0.11
## [13] robustbase_0.93-5 mzID_1.24.0 labeling_0.3
## [16] GenomeInfoDbData_1.2.2 farver_2.0.3 bit64_0.9-7
## [19] rprojroot_1.3-2 vctrs_0.2.4 xfun_0.12
## [22] BiocFileCache_1.10.2 R6_2.4.1 doParallel_1.0.15
## [25] ggbeeswarm_0.6.0 clue_0.3-57 rsvd_1.0.3
## [28] locfit_1.5-9.1 bitops_1.0-6 assertthat_0.2.1
## [31] nnet_7.3-13 beeswarm_0.2.3 gtable_0.3.0
## [34] affy_1.64.0 rlang_0.4.5 GlobalOptions_0.1.1
## [37] splines_3.6.1 acepack_1.4.1 impute_1.60.0
## [40] checkmate_2.0.0 BiocManager_1.30.10 yaml_2.2.1
## [43] reshape2_1.4.3 backports_1.1.5 Hmisc_4.3-1
## [46] MassSpecWavelet_1.52.0 RBGL_1.62.1 tools_3.6.1
## [49] ellipsis_0.3.0 affyio_1.56.0 gplots_3.0.3
## [52] RColorBrewer_1.1-2 ggridges_0.5.2 plyr_1.8.6
## [55] base64enc_0.1-3 progress_1.2.2 zlibbioc_1.32.0
## [58] RCurl_1.98-1.1 prettyunits_1.1.1 rpart_4.1-15
## [61] GetoptLong_0.1.8 viridis_0.5.1 cluster_2.1.0
## [64] colorRamps_2.3 data.table_1.12.8 circlize_0.4.8
## [67] RANN_2.6.1 pcaMethods_1.78.0 packrat_0.5.0
## [70] hms_0.5.3 evaluate_0.14 pbkrtest_0.4-8.6
## [73] XML_3.99-0.3 jpeg_0.1-8.1 shape_1.4.4
## [76] compiler_3.6.1 KernSmooth_2.23-16 ncdf4_1.17
## [79] crayon_1.3.4 minqa_1.2.4 htmltools_0.4.0
## [82] mgcv_1.8-31 Formula_1.2-3 lubridate_1.7.4
## [85] DBI_1.1.0 dbplyr_1.4.2 MASS_7.3-51.5
## [88] rappdirs_0.3.1 boot_1.3-24 Matrix_1.2-18
## [91] cli_2.0.2 vsn_3.54.0 gdata_2.18.0
## [94] igraph_1.2.4.2 pkgconfig_2.0.3 foreign_0.8-76
## [97] MALDIquant_1.19.3 vipor_0.4.5 dqrng_0.2.1
## [100] multtest_2.42.0 XVector_0.26.0 digest_0.6.25
## [103] graph_1.64.0 rmarkdown_2.1 htmlTable_1.13.3
## [106] edgeR_3.28.1 DelayedMatrixStats_1.8.0 listarrays_0.3.1
## [109] curl_4.3 gtools_3.8.1 rjson_0.2.20
## [112] nloptr_1.2.2.1 lifecycle_0.2.0 nlme_3.1-145
## [115] BiocNeighbors_1.4.2 fansi_0.4.1 viridisLite_0.3.0
## [118] pillar_1.4.3 lattice_0.20-40 httr_1.4.1
## [121] DEoptimR_1.0-8 survival_3.1-11 glue_1.3.1
## [124] png_0.1-7 iterators_1.0.12 bit_1.1-15.2
## [127] stringi_1.4.6 blob_1.2.1 BiocSingular_1.2.2
## [130] latticeExtra_0.6-29 caTools_1.18.0 memoise_1.1.0
## [133] irlba_2.3.3