Visualize simulated batch effects
Almut Lütge
14 May, 2020
hca
suppressPackageStartupMessages({
library(scater)
library(CellMixS)
library(purrr)
library(data.table)
library(here)
library(tidyr)
library(dplyr)
library(stringr)
library(ComplexHeatmap)
library(edgeR)
library(scran)
library(cowplot)
library(ggrepel)
library(readr)
})Dataset and parameter
sce <- readRDS(params$data)
sce_real <- readRDS(params$real)
sim_list = list(strsplit(params$sim, ","))
print(sim_list[[1]][[1]])## [1] "1__1_0" "2__1_0" "0.9__2_1" "0.5__1" "4__1" "0.7__1_0"
## [7] "2__2_1" "0.7__2_1" "0.5__2_1" "0.5__1_0" "0.3__1" "1.2__1"
## [13] "0.8__1_0" "0.6__1_0" "1__1" "1.2__2_1" "0.8__2_1" "4__1_0"
## [19] "0.3__1_0" "0.4__2_1" "0__0" "0.8__1" "2__1" "1.5__2_1"
## [25] "0.9__1_0" "4__2_1" "1.2__1_0" "1.5__1_0" "0.6__2_1" "0.4__1_0"
## [31] "1.5__1" "1__2_1" "0.7__1" "0.6__1" "0.9__1" "0.3__2_1"
## [37] "0.4__1"param <- readRDS(params$param)
celltype <- param[["celltype"]]
batch <- param[["batch"]]
sample <- param[["Sample"]]
dataset_name <- param[["dataset_name"]]
param_real <- readRDS(params$param_real)
batch_real <- param_real[["batch"]]
cluster_real <- param_real[["celltype"]]
sample_real <- param_real[["sample"]]
table(colData(sce_real)[,sample_real])##
## C1HT-medium C1HT-small CEL-Seq2 Chromium Chromium(sn) ddSEQ
## 2210 1603 1082 1604 1512 2104
## Drop-Seq ICELL8 inDrop MARS-Seq mcSCRB-Seq Quartz-Seq2
## 2260 1912 685 1476 1680 1332
## Smart-Seq2
## 732#colors for plotting
tsne_cols <- c(
"#DC050C", "#1965B0", "#FB8072","#7BAFDE", "#B17BA6",
"#FDB462", "#882E72", "#FF7F00", "#E78AC3", "#B2DF8A",
"#33A02C", "#55A1B1", "#B2DF8A", "#8DD3C7", "#A6761D",
"#7570B3", "#E6AB02", "#BEAED4", "#666666", "#999999")
mds_cols <- c(
"#DC050C", "#1965B0", "#FB8072","#7BAFDE", "#B17BA6",
"#FDB462", "#882E72", "#FF7F00", "#E78AC3", "#B2DF8A",
"#33A02C", "#55A1B1", "#B2DF8A", "#8DD3C7", "#A6761D",
"#7570B3", "#E6AB02", "#BEAED4", "#666666", "#999999")Visualize simulations
#Plot tsne
visUMAP <- function(simulation, title, ids){
p <- visGroup(simulation, ids, dim_red = "UMAP")
p[["data"]]$cluster_id <- colData(simulation)[, "cluster_id"]
p + aes(shape = cluster_id) +
scale_shape_manual(values=1:nlevels(simulation$cluster_id)) +
guides(colour = guide_legend(override.aes = list(size=3, alpha = 1))) +
guides(shape = guide_legend(override.aes = list(size=3, alpha = 1), title = "celltype")) +
geom_point(size=2, alpha = 1, aes_string(color="group_var")) +
scale_color_manual(values = tsne_cols) +
ggtitle(title)
}
visTSNE <- function(simulation, title, ids){
p <- visGroup(simulation, ids)
p[["data"]]$cluster_id <- colData(simulation)[, "cluster_id"]
p + aes(shape = cluster_id) +
scale_shape_manual(values=1:nlevels(simulation$cluster_id)) +
guides(colour = guide_legend(override.aes = list(size=3, alpha = 1))) +
guides(shape = guide_legend(override.aes = list(size=3, alpha = 1), title = "celltype")) +
geom_point(size=2, alpha = 1, aes_string(color="group_var")) +
scale_color_manual(values = tsne_cols) +
ggtitle(title)
}
#Functions from muscat to generate pseudobulk sample and genrate a MDSplot
pbMDS <- function(x, title) {
y <- as(assays(x), "list")
y <- do.call("cbind", y)
d <- suppressMessages(DGEList(y))
d <- calcNormFactors(d)
mds <- plotMDS.DGEList(d, plot = FALSE)
ei <- metadata(x)$experiment_info
m <- match(colnames(x), ei$sample_id)
nk <- length(assays(x))
df <- data.frame(
MDS1 = mds$x,
MDS2 = mds$y,
cluster_id = rep(assayNames(x), each = ncol(x)),
group_id = rep(ei$group_id[m], nk),
sample_id = colnames(x))
cols <- mds_cols
if (nk > length(cols))
cols <- colorRampPalette(cols)(nk)
ggplot(df, aes_string(x = "MDS1", y = "MDS2",
col = "sample_id", shape = "cluster_id")) +
scale_color_manual(values = cols) +
scale_shape_manual(values=1:nlevels(df$cluster_id)) +
geom_point(size = 5, alpha = .8) +
guides(color = guide_legend(override.aes = list(alpha = 1))) +
ggtitle(title) +
theme_bw() + theme(aspect.ratio = 1,
axis.text = element_text(color = "black"),
panel.grid.minor = element_blank(),
panel.grid.major = element_line(size = 0.2, color = "lightgrey"))
}
#helper functions from muscat
# Compute pseudo bulks
.pb <- function(x, cs, assay, fun) {
fun <- switch(fun,
rowMedians = getFromNamespace(fun, "matrixStats"),
getFromNamespace(fun, "Matrix"))
pb <- map_depth(cs, -1, function(i) {
if (length(i) == 0) return(numeric(nrow(x)))
fun(assays(x)[[assay]][, i, drop = FALSE])
})
map_depth(pb, -2, function(u)
data.frame(u,
row.names = rownames(x),
check.names = FALSE))
}
#split cells
.split_cells <- function(x,
by = c("cluster_id", "sample_id")) {
if (is(x, "SingleCellExperiment"))
x <- colData(x)
cd <- data.frame(x[by], check.names = FALSE)
cd <- data.table(cd, cell = rownames(cd)) %>%
split(by = by, sorted = TRUE, flatten = FALSE)
map_depth(cd, length(by), "cell")
}
aggregateData <- function(x, assay,
by = c("cluster_id", "sample_id"),
fun = c("sum", "mean", "median"),
scale = FALSE) {
if (missing("assay"))
assay <- assayNames(x)[1]
# validity checks for input arguments
stopifnot(is.character(by), by %in% colnames(colData(x)))
stopifnot(is.logical(scale), length(scale) == 1)
# get aggregation function
fun <- match.arg(fun)
fun <- switch(fun,
sum = "rowSums",
mean = "rowMeans",
median = "rowMedians")
# split cells & compute pseudo-bulks
cs <- .split_cells(x, by)
pb <- .pb(x, cs, assay, fun)
# scale
if (scale) {
if (assay == "counts" & fun == "rowSums") {
pb_counts <- pb
} else {
pb_counts <- .pb(x, cs, assay, "rowSums")
}
pb <- map_depth(pb_counts, -2, function(u)
u / colSums(u)[col(u)] * 1e6)
}
pb <- map_depth(pb, -2, as.matrix)
# return SCE
md <- metadata(x)
md$agg_pars <- list(assay = assay, fun = fun)
SingleCellExperiment(
assays = pb,
metadata = md)
}
visMDS <-function(simulation, title, ids){
agg_sim <- aggregateData(simulation, by = c("cluster_id", ids),
fun = "mean", scale = TRUE)
pbMDS(agg_sim, title)
}TSNE
1__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.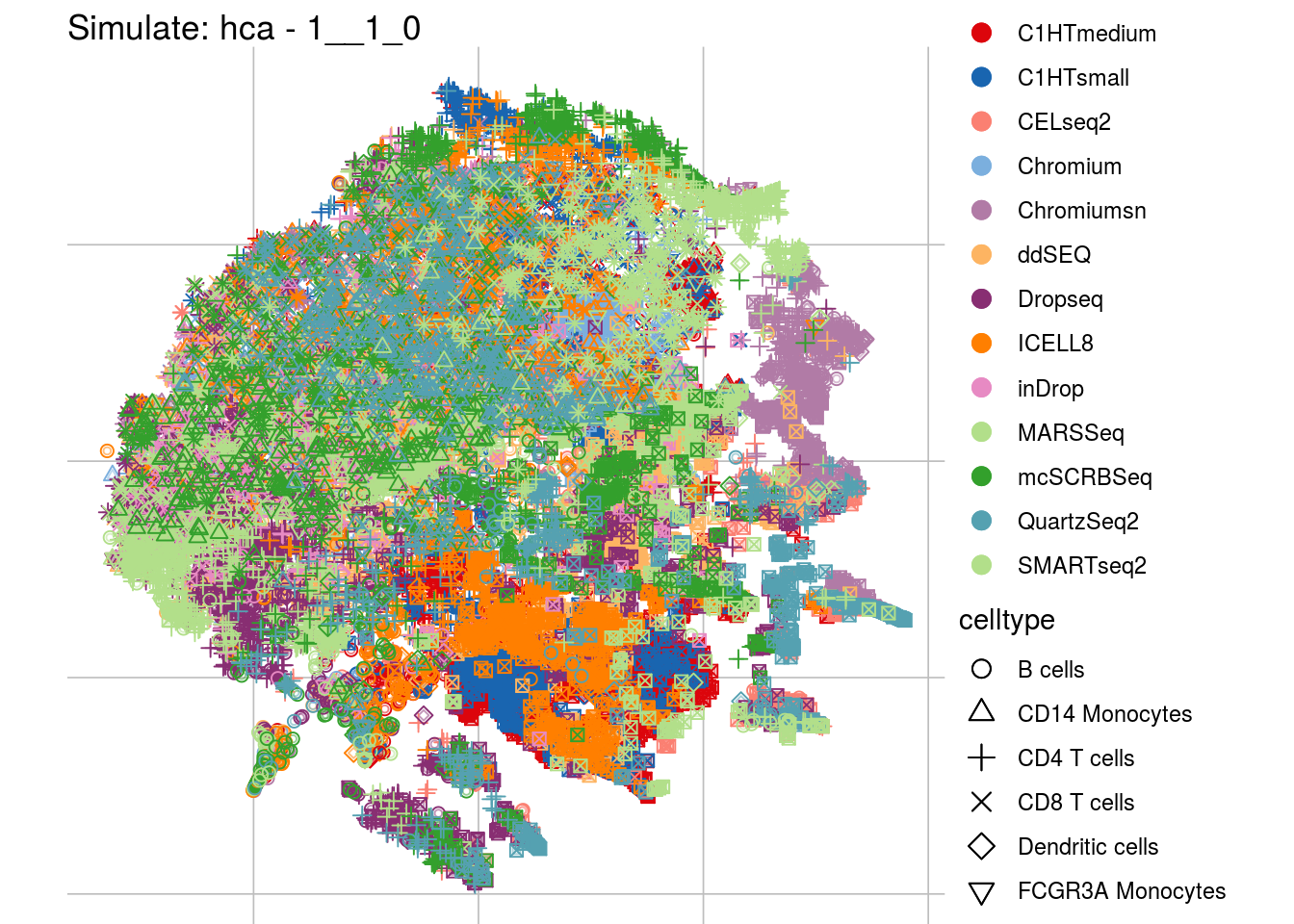
2__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.9__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.5__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
4__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.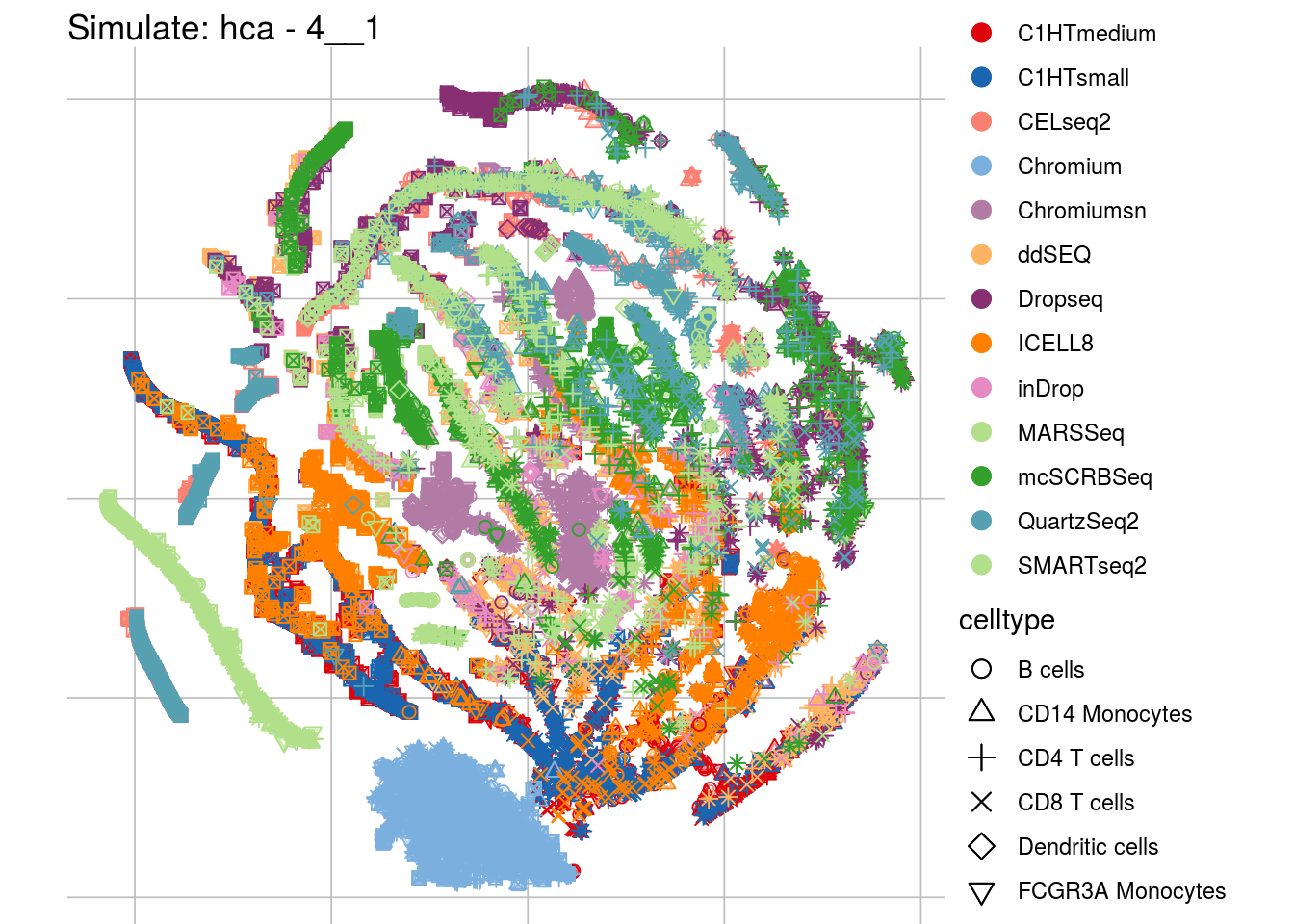
0.7__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
2__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.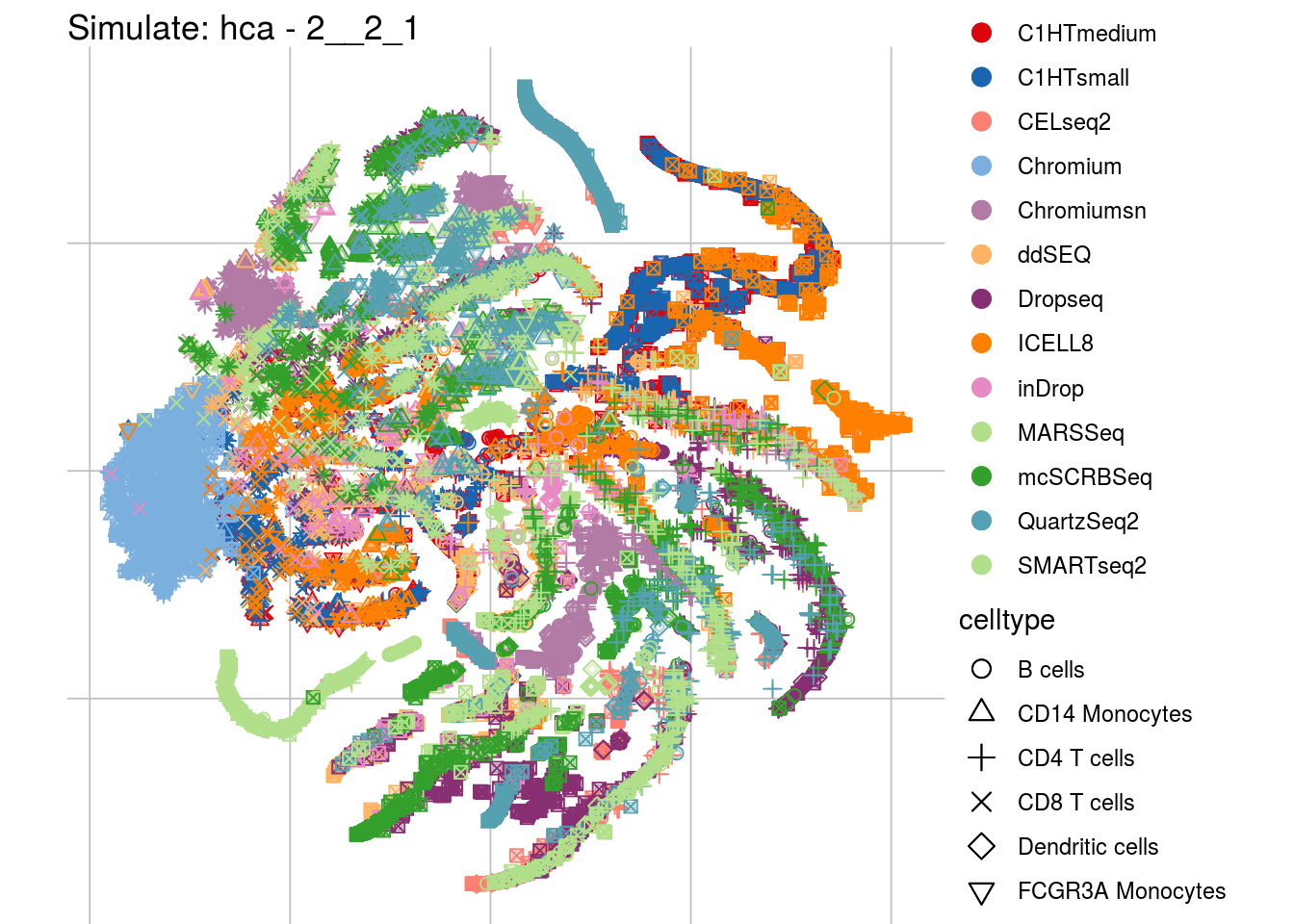
0.7__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.5__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.5__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.3__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
1.2__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.8__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.6__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
1__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
1.2__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.8__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
4__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.3__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.4__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.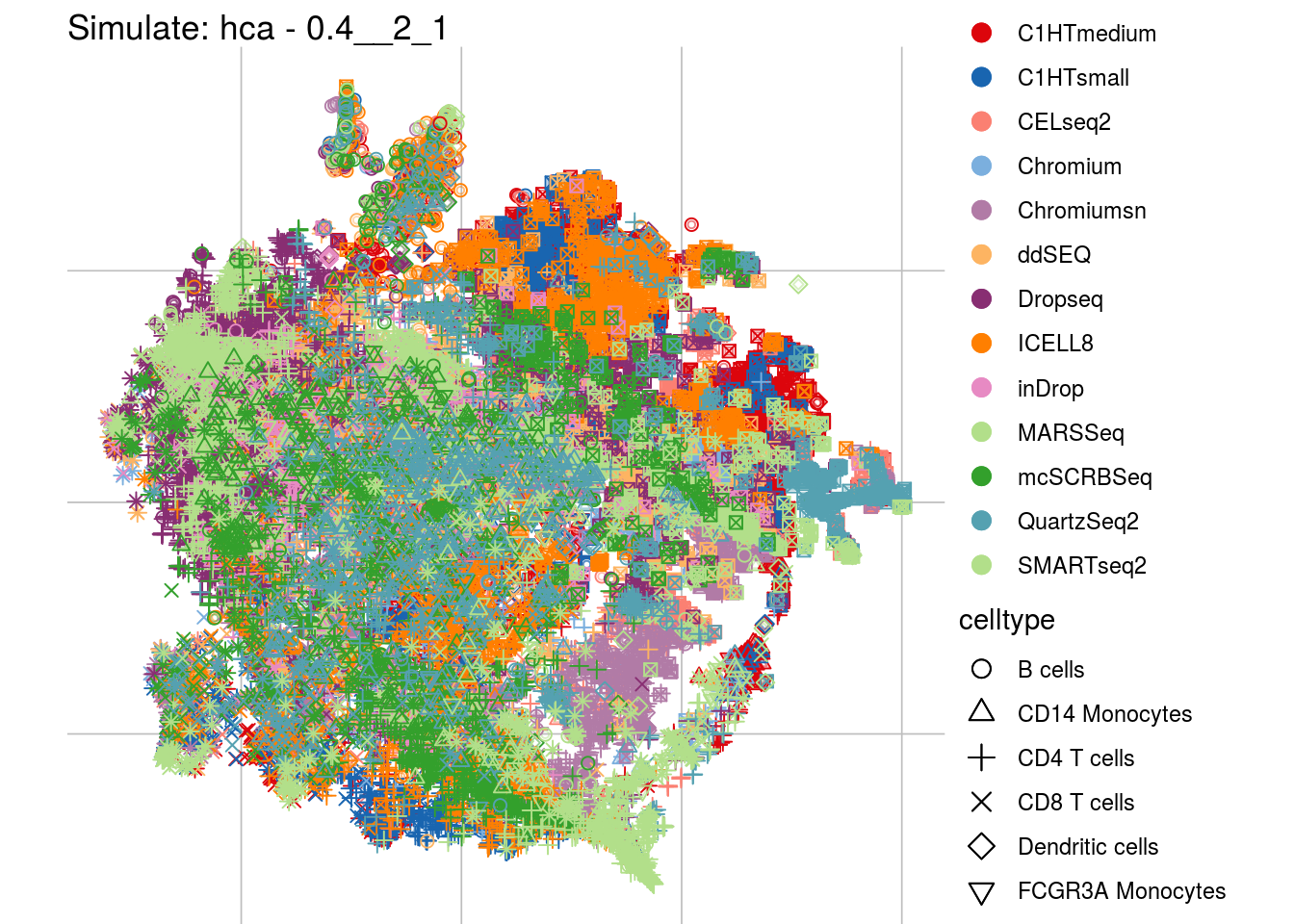
0__0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.8__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.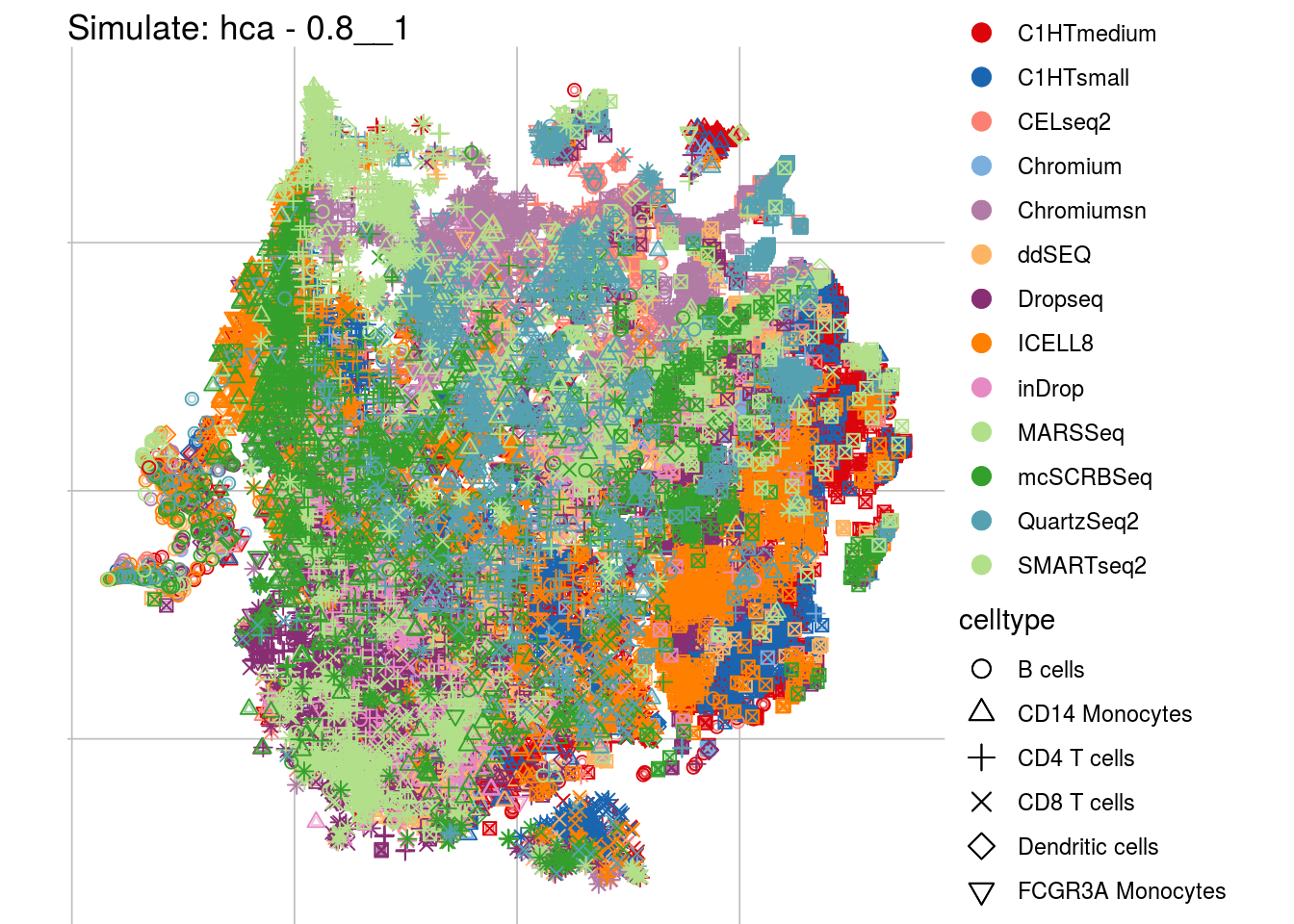
2__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.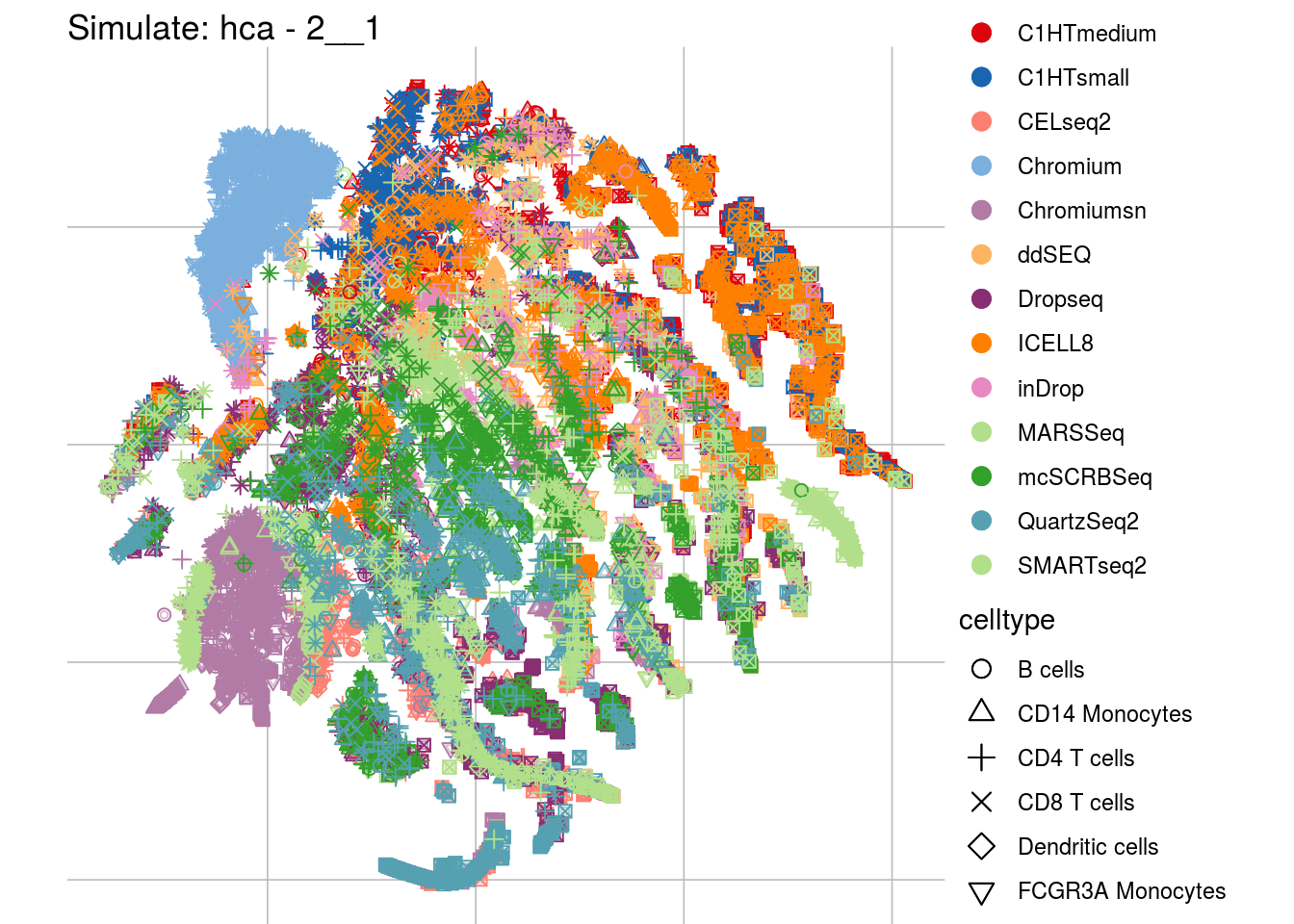
1.5__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.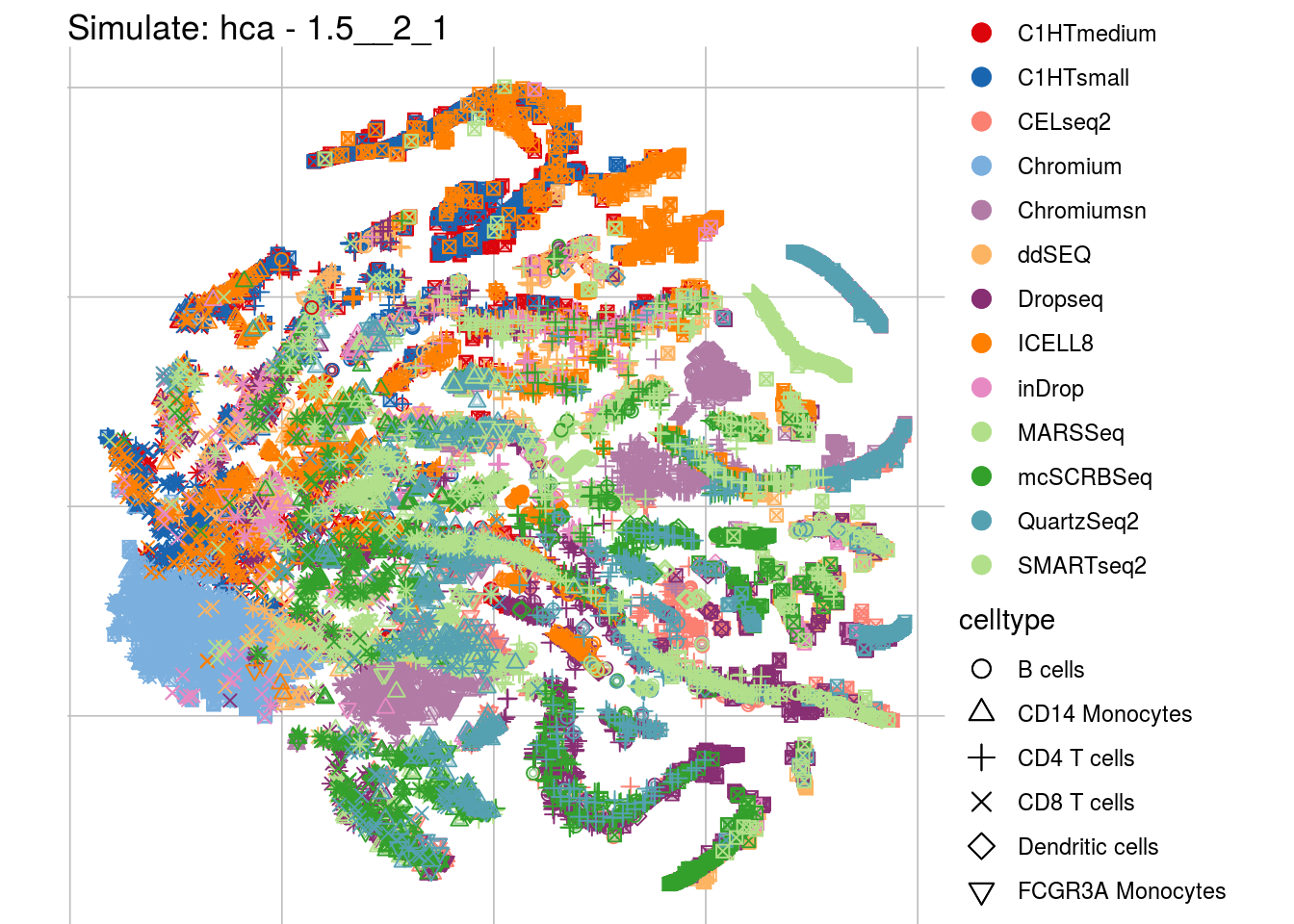
0.9__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
4__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.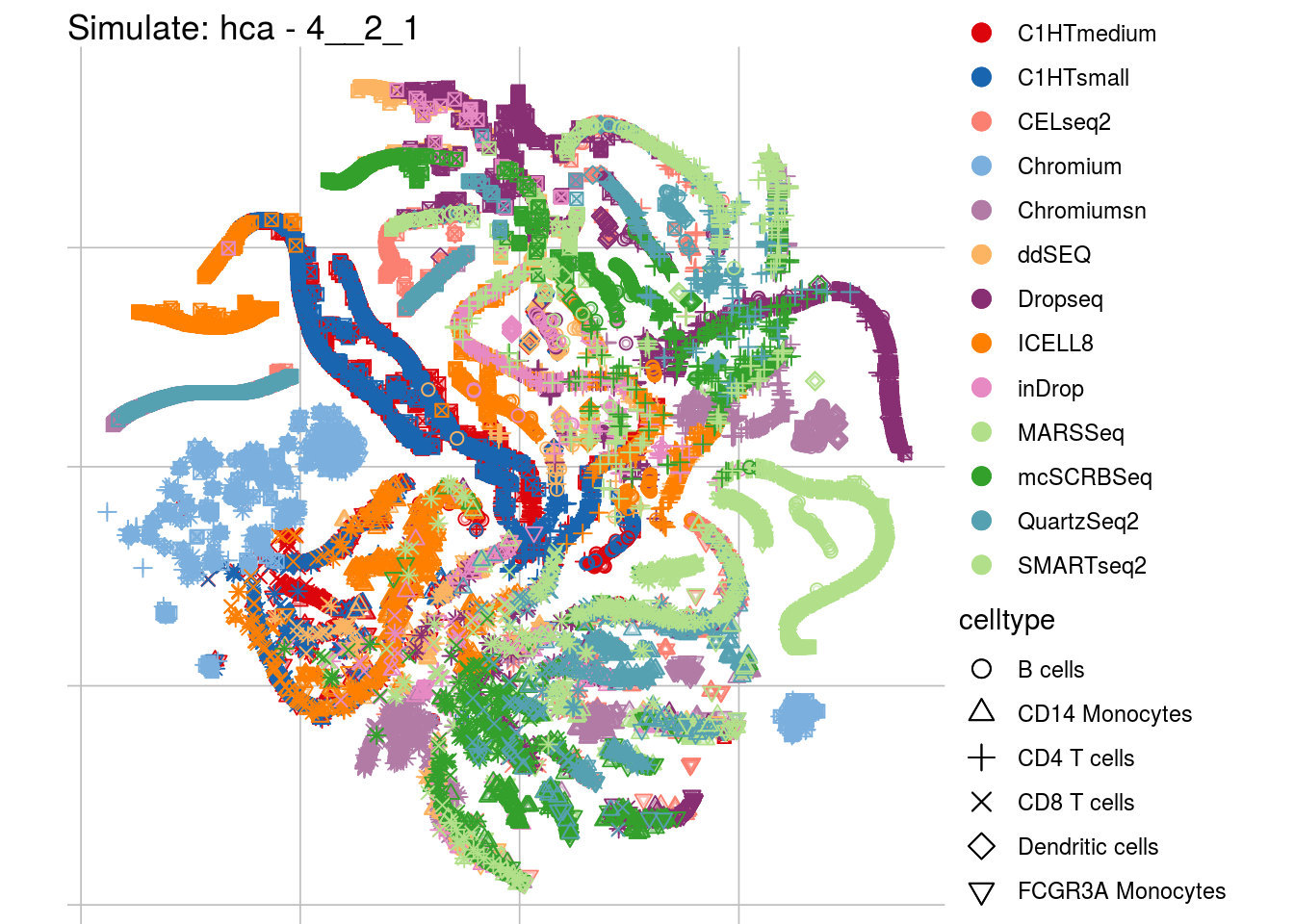
1.2__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.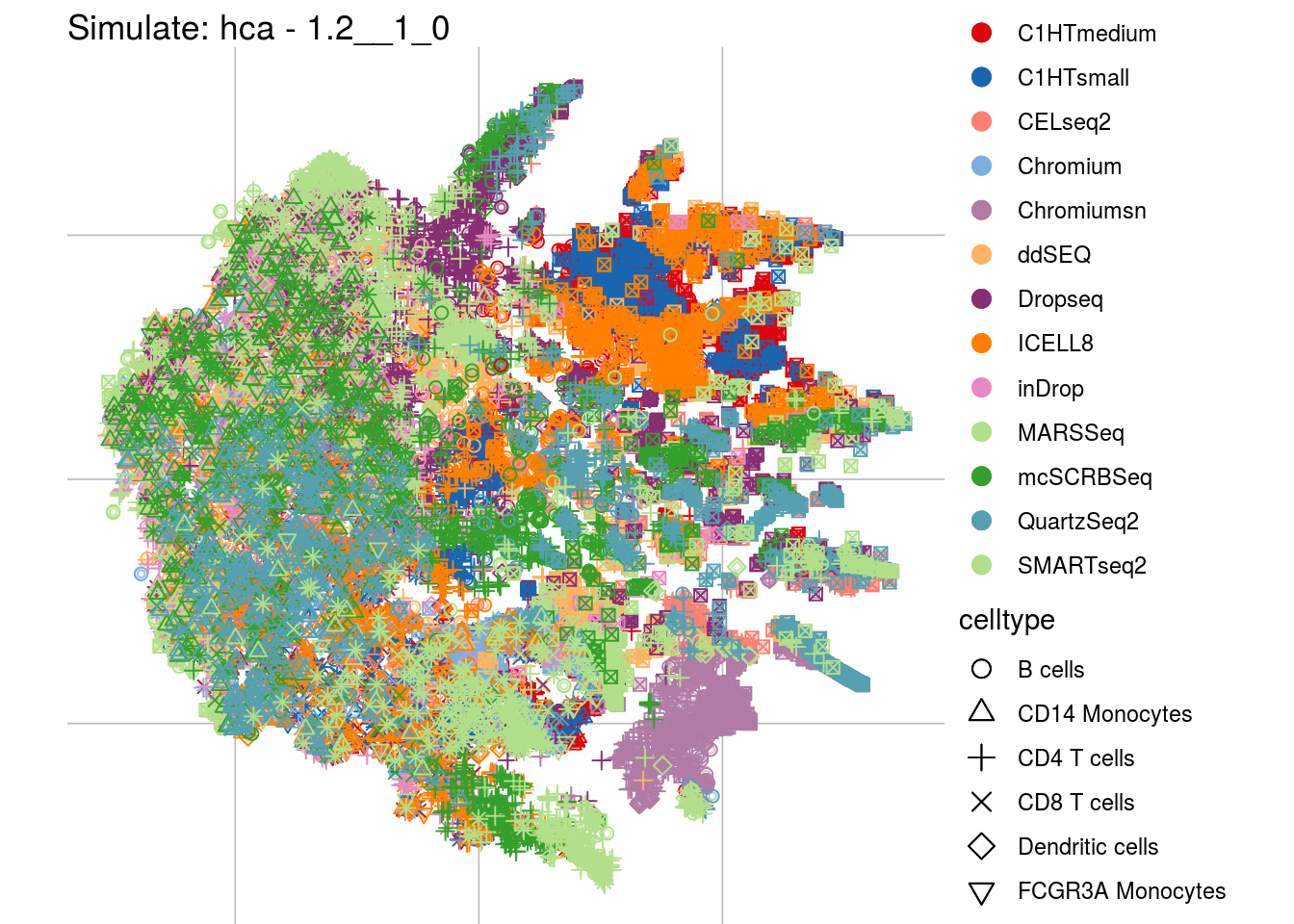
1.5__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.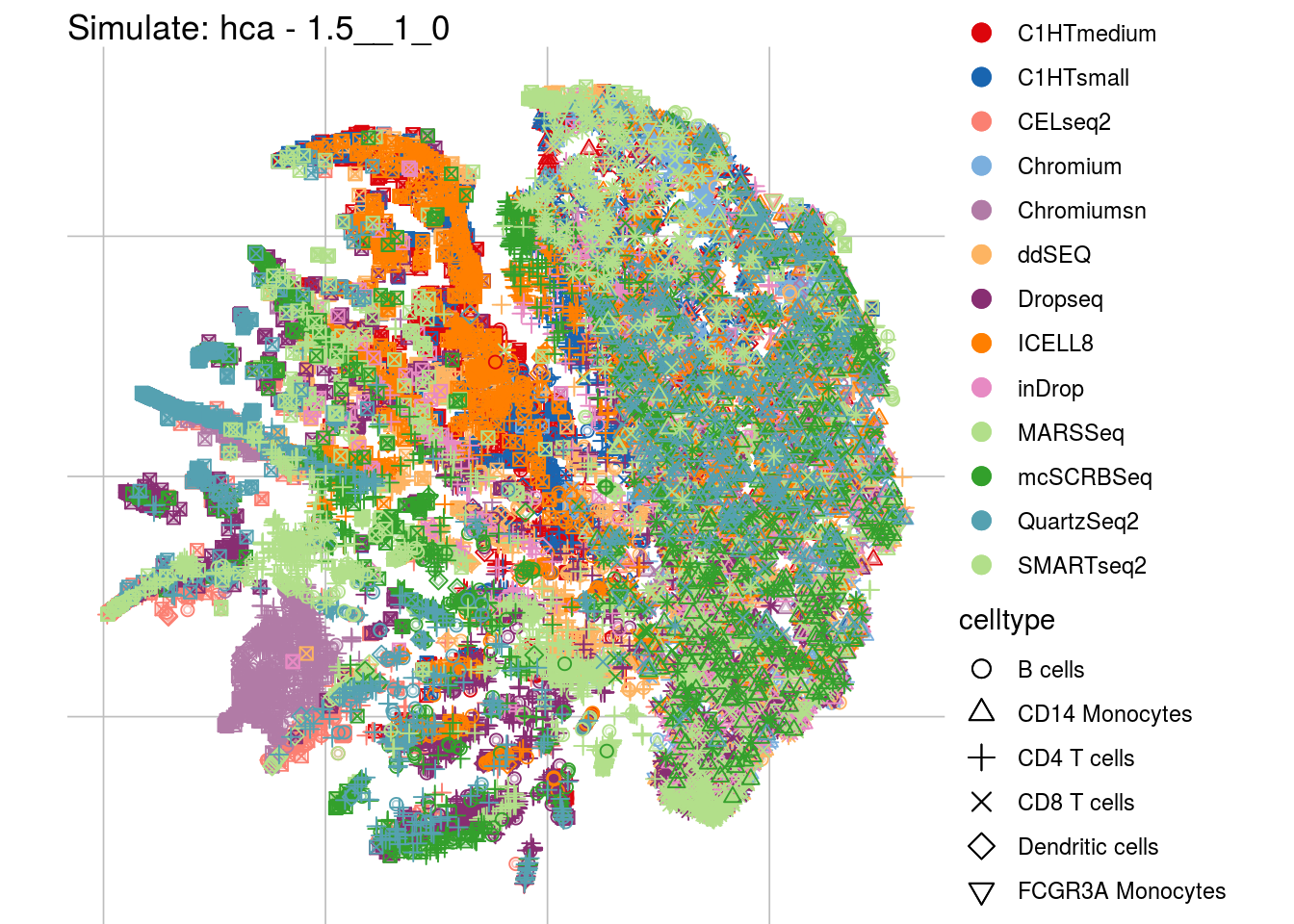
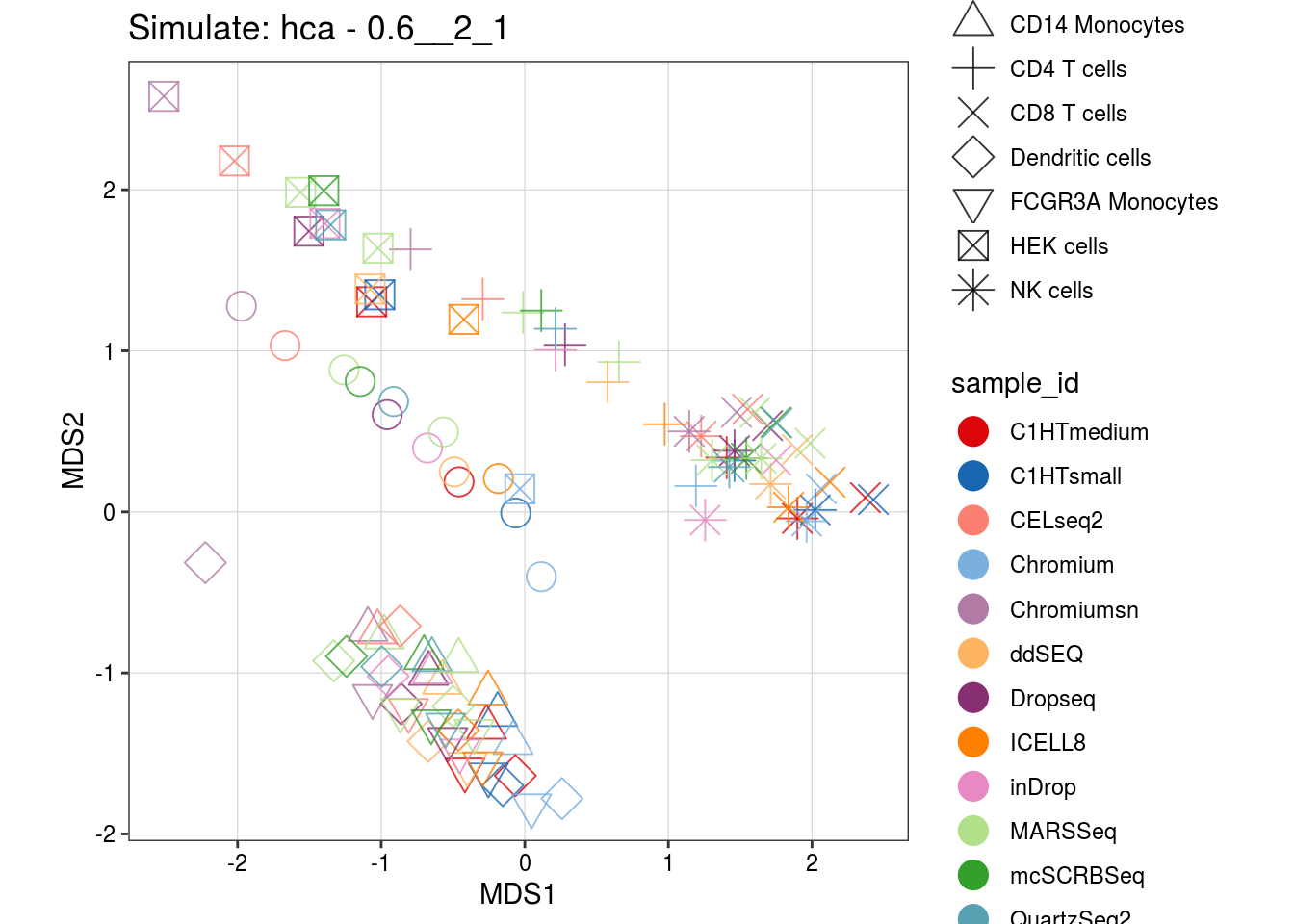
0.6__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
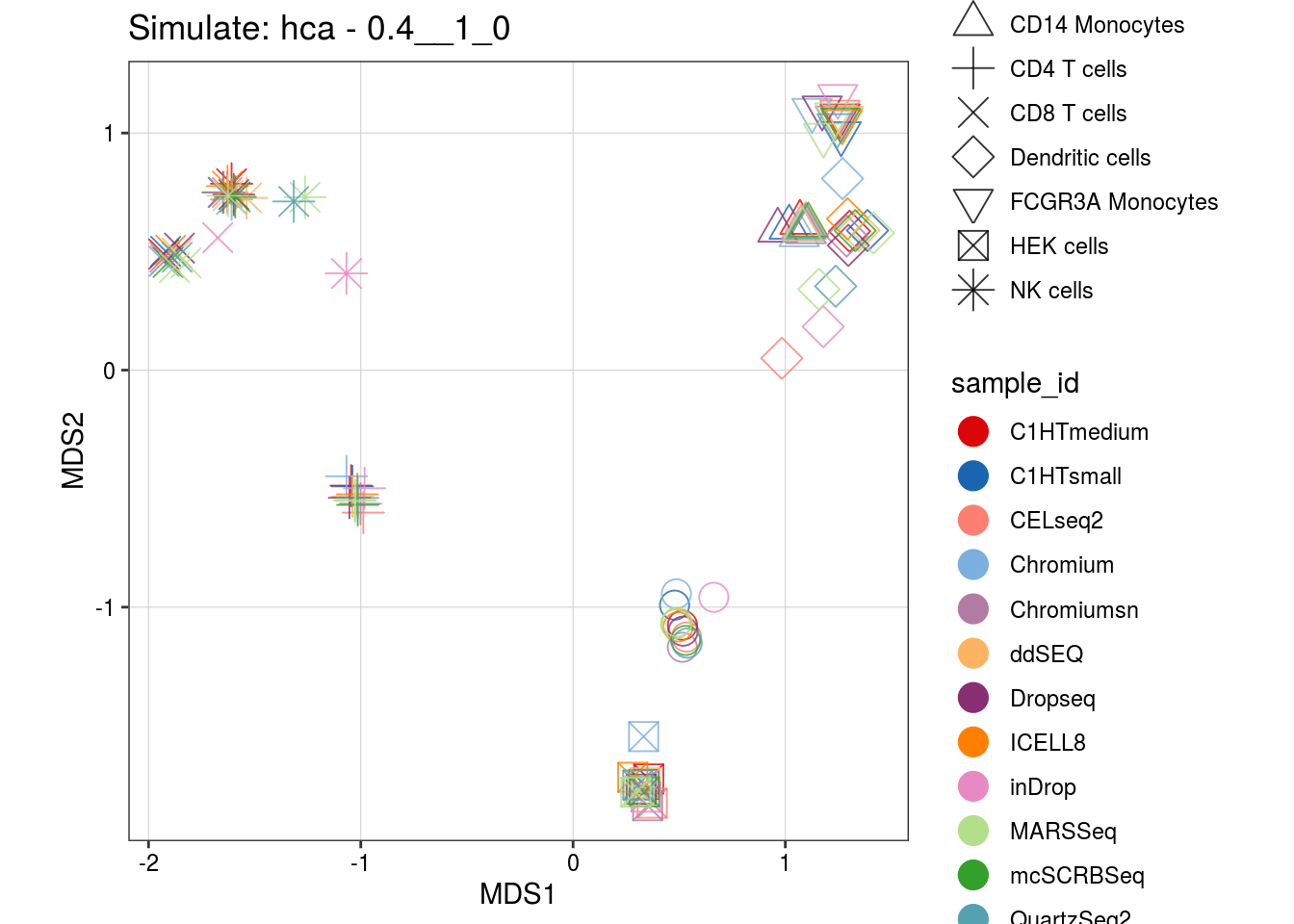
0.4__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.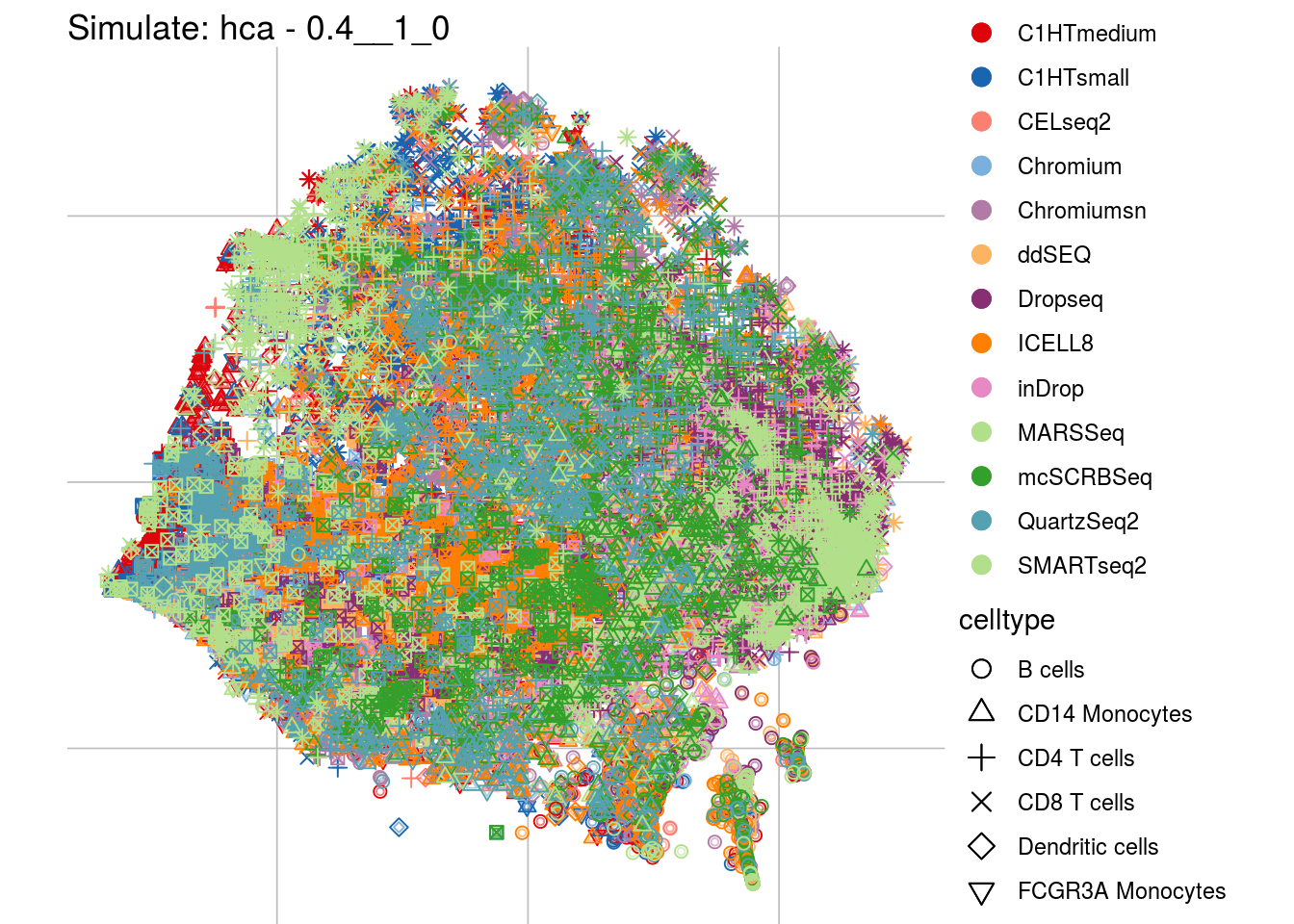
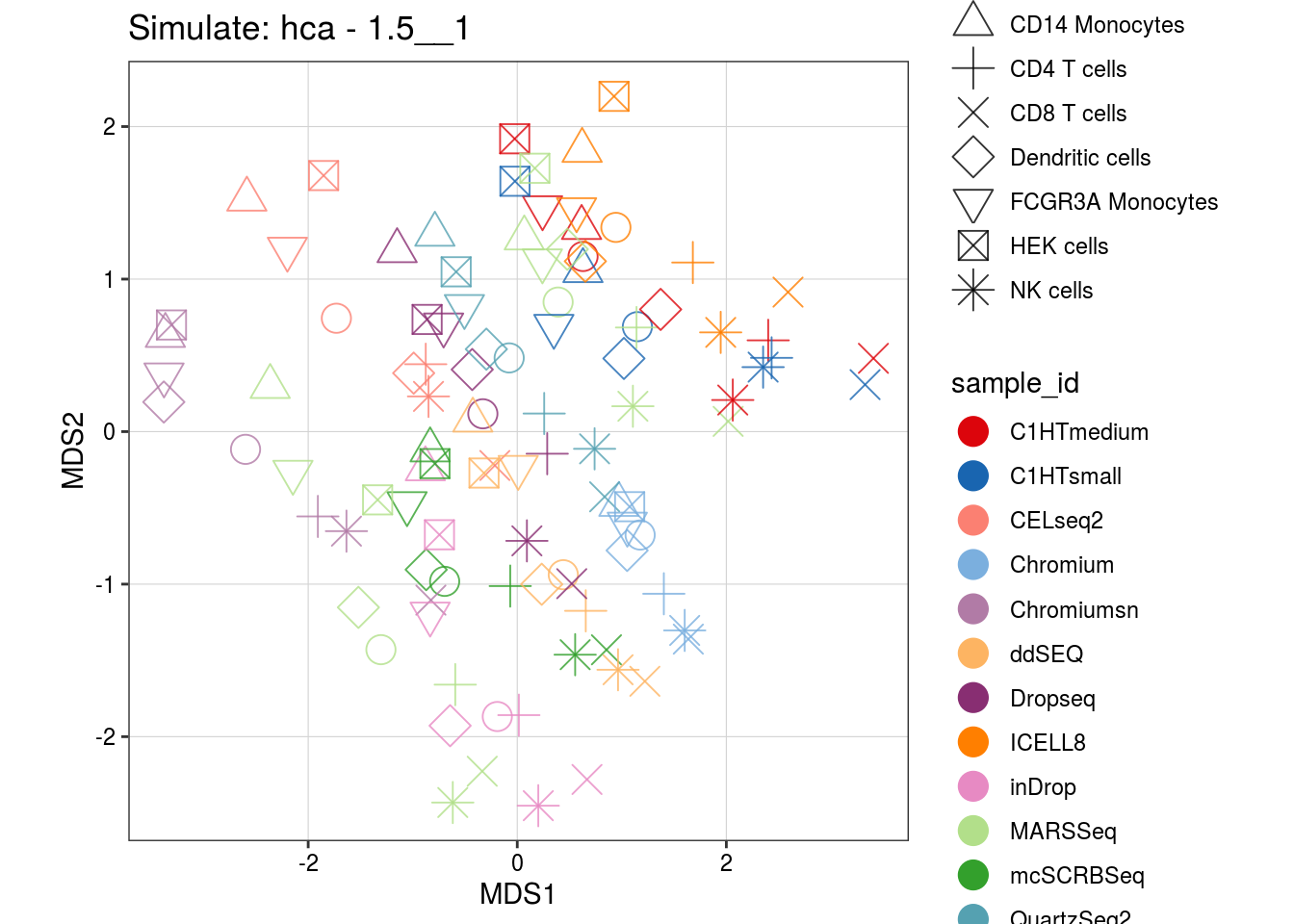
1.5__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
1__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.7__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
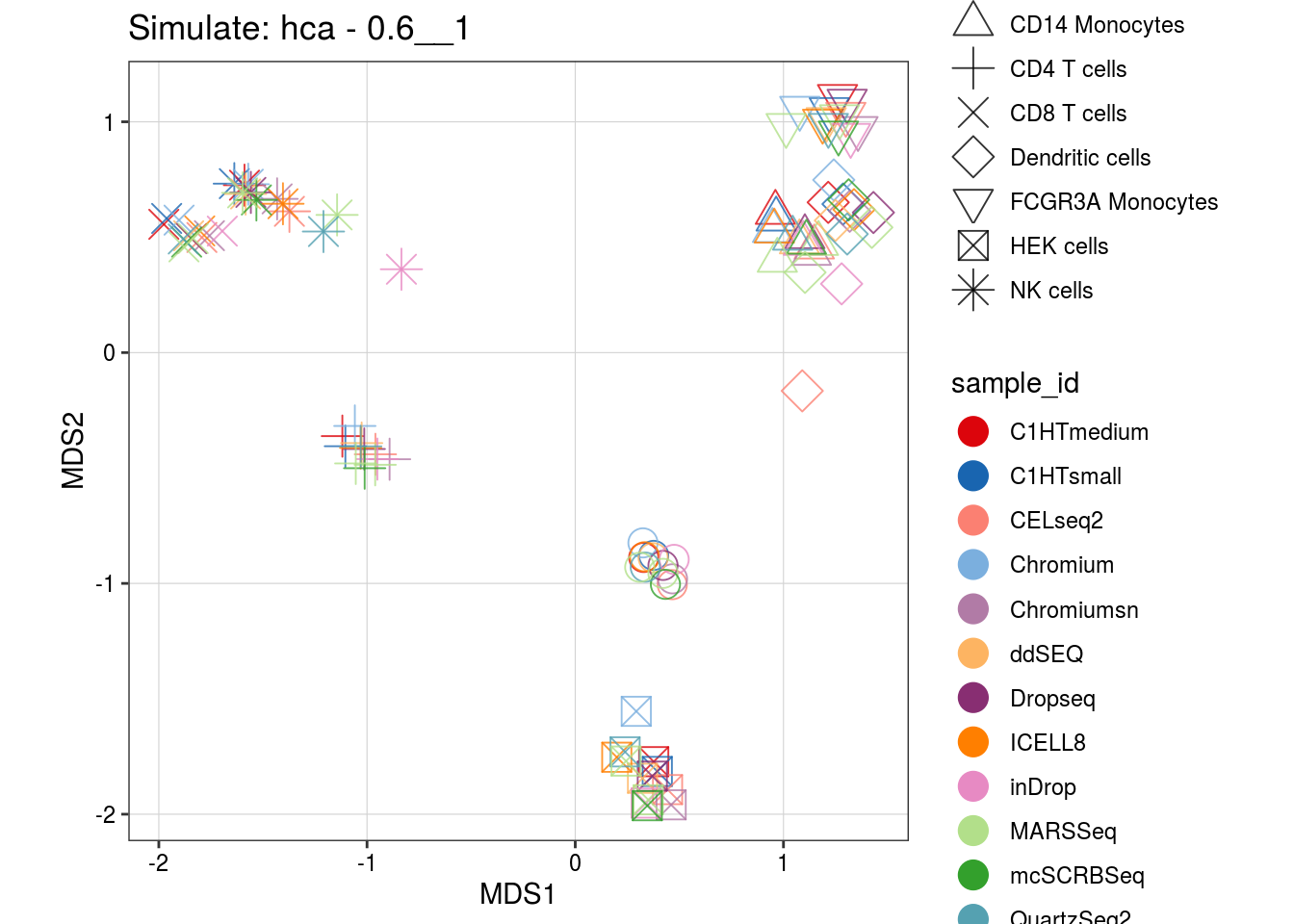
0.6__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.9__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.3__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.4__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
UMAP
template_umap <- c(
"#### {{nm}}\n",
"```{r umap{{nm}}, echo = FALSE}\n",
"filename <- paste0('out/sim_char/', '{{name}}', '/sim_', '{{name}}', '_', '{{nm}}', '_sce.rds')",
"sim_sce <- readRDS(file = filename)",
"visUMAP(sim_sce, title = paste0('Simulate: ', '{{name}}', ' - ', '{{nm}}'), ids = batch)\n",
"```\n",
"\n"
)
plots_umap <- lapply(sim_list[[1]][[1]],
function(nm, name = dataset_name) knitr::knit_expand(text = template_umap)
)1__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
2__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.9__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.5__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
4__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.7__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
2__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.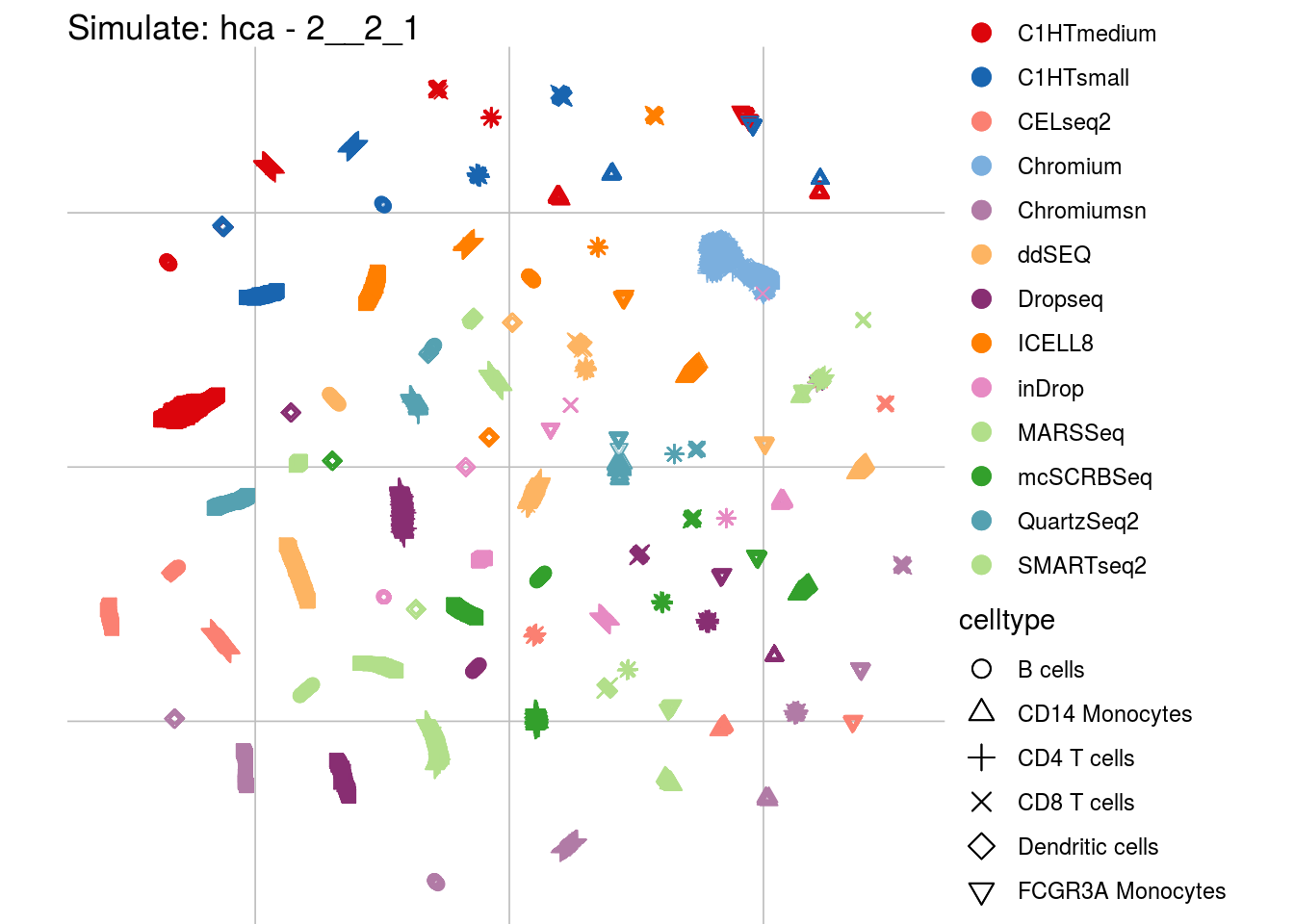
0.7__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.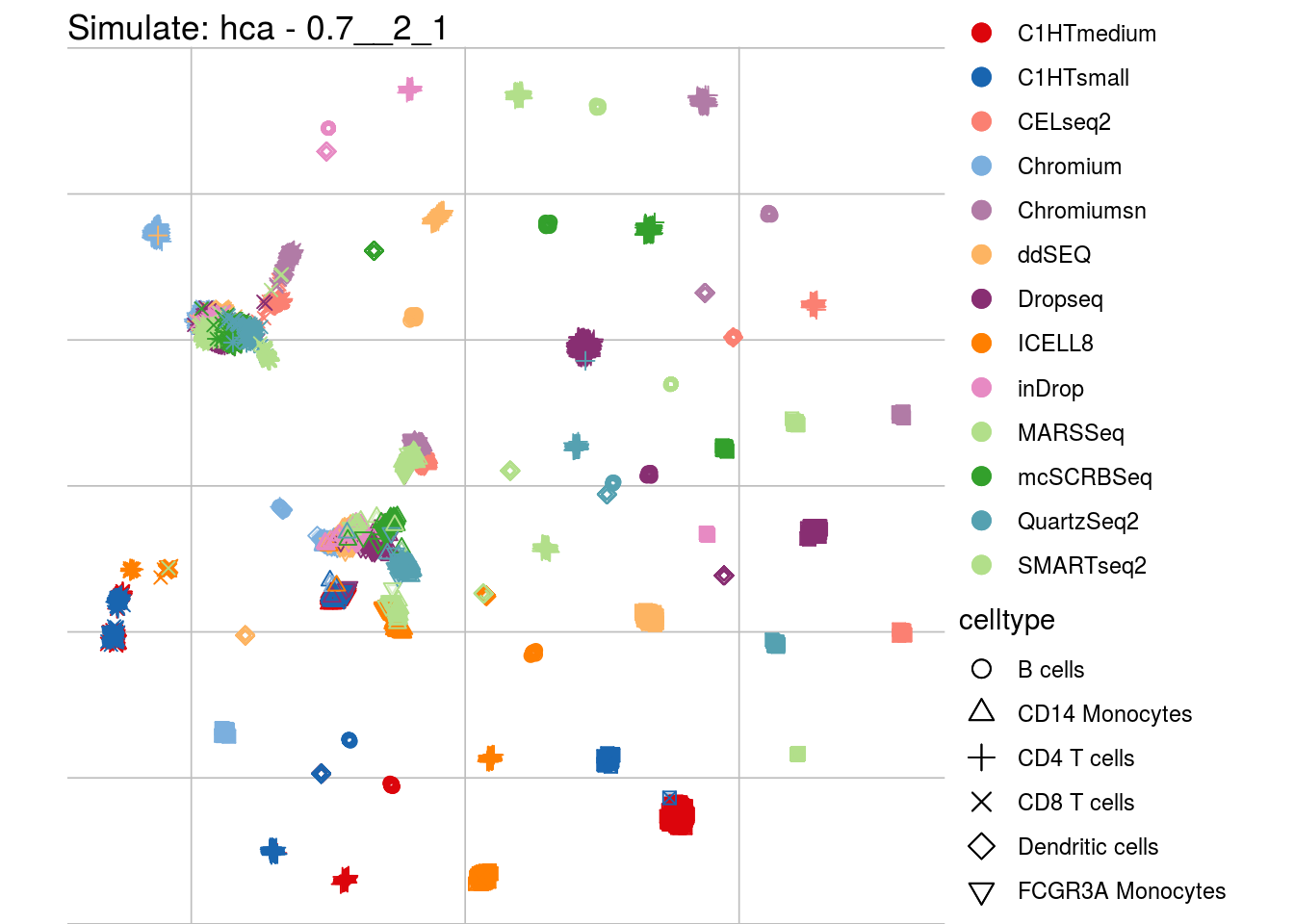
0.5__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.5__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.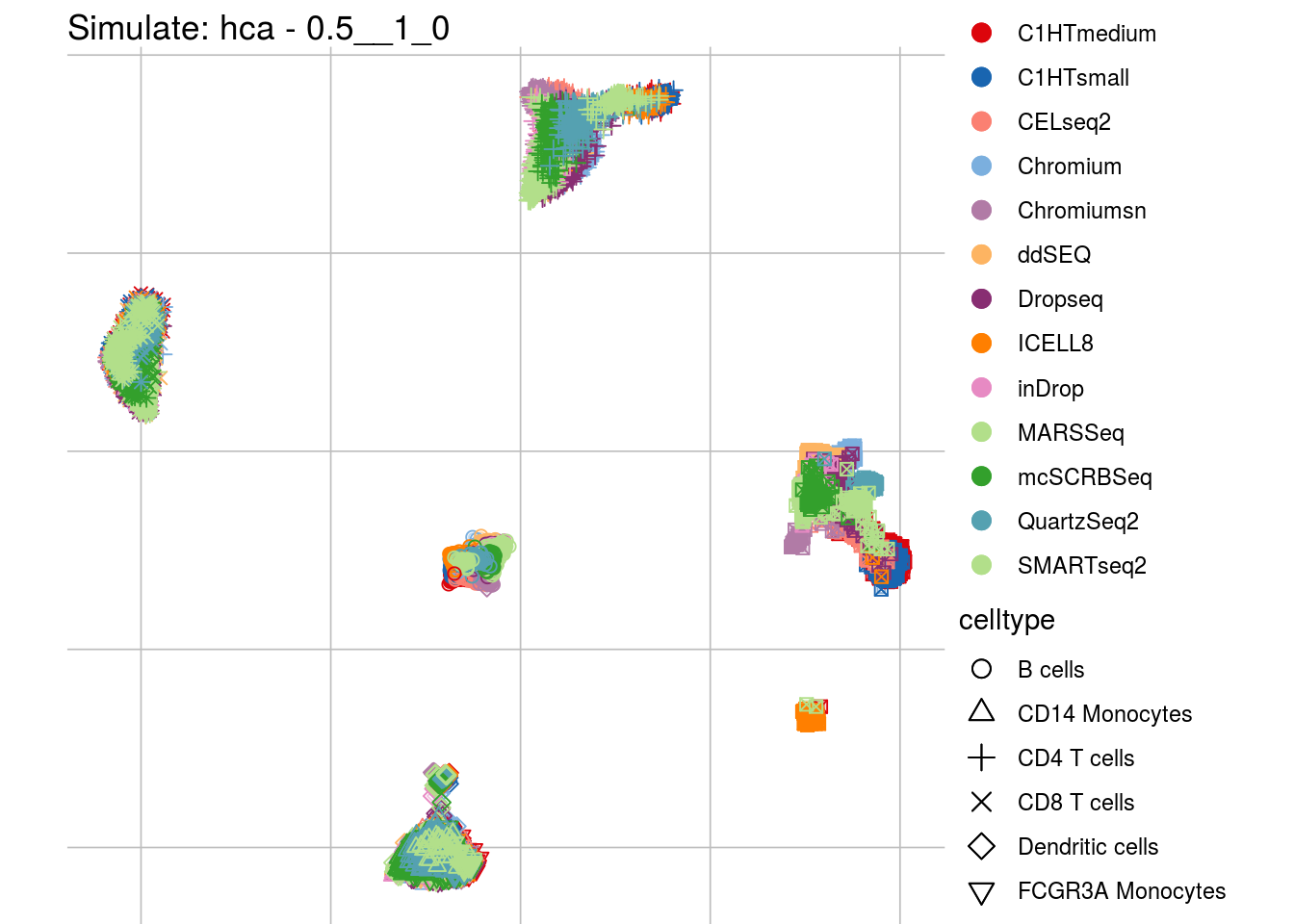
0.3__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
1.2__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.8__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.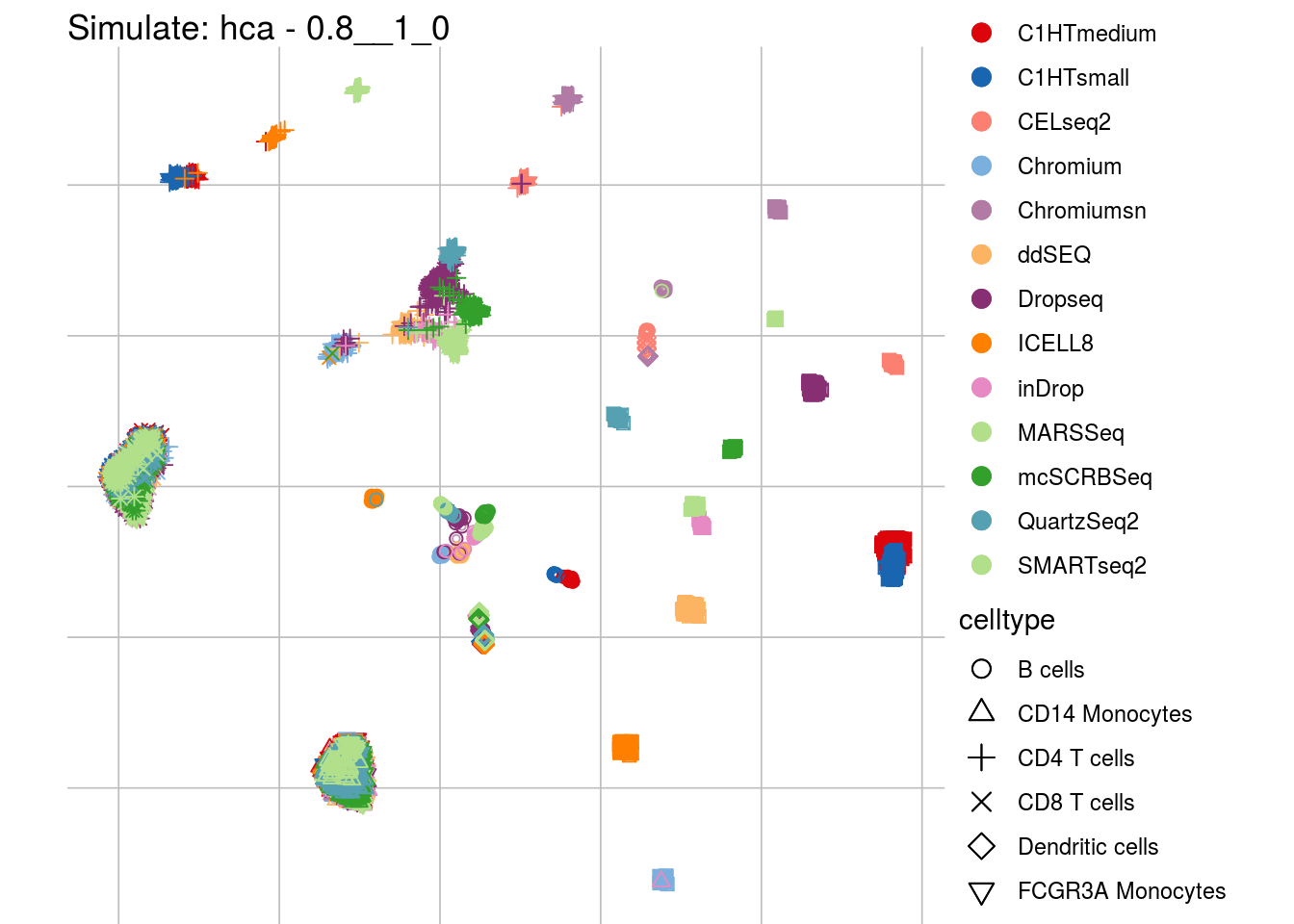
0.6__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
1__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.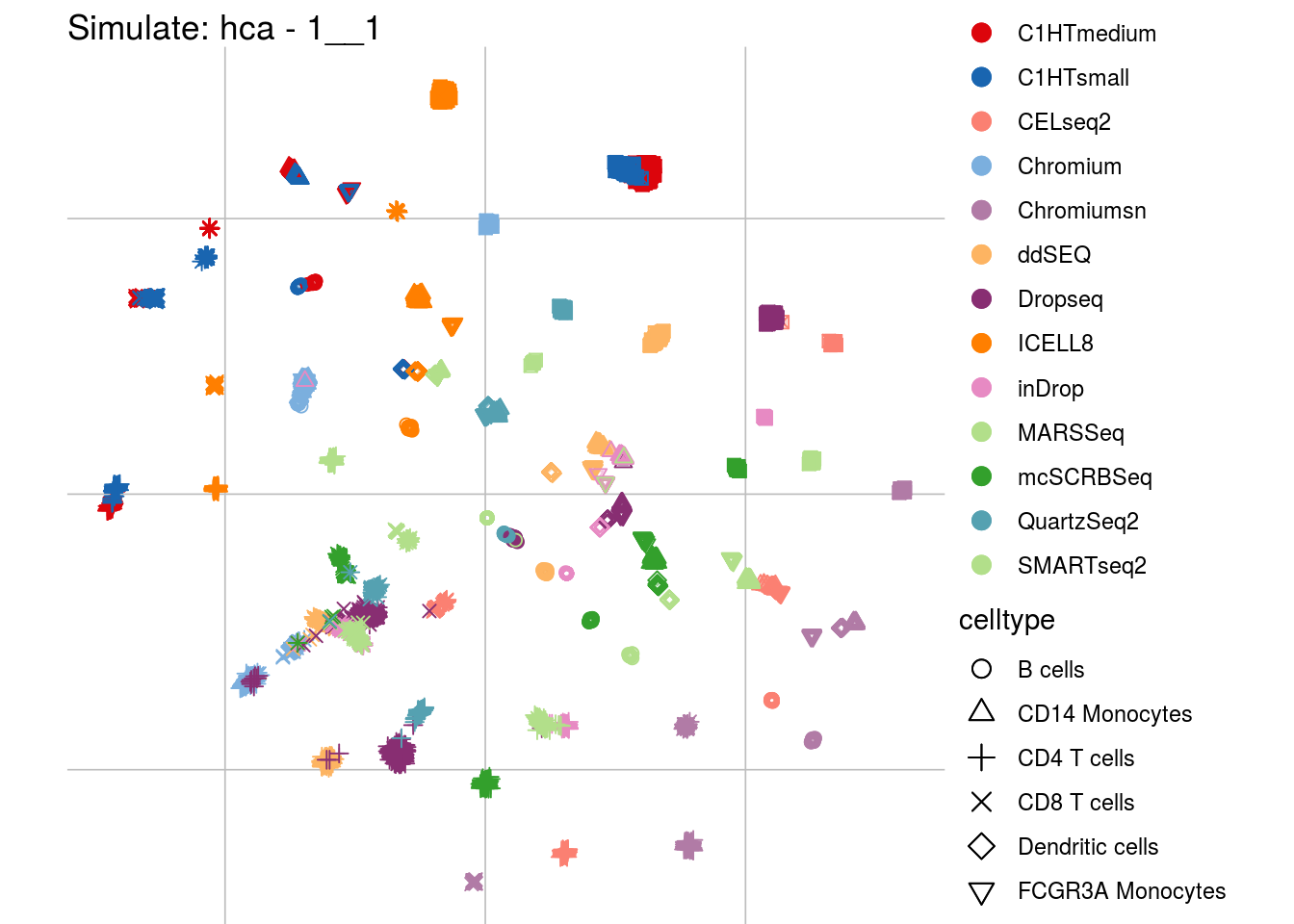
1.2__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.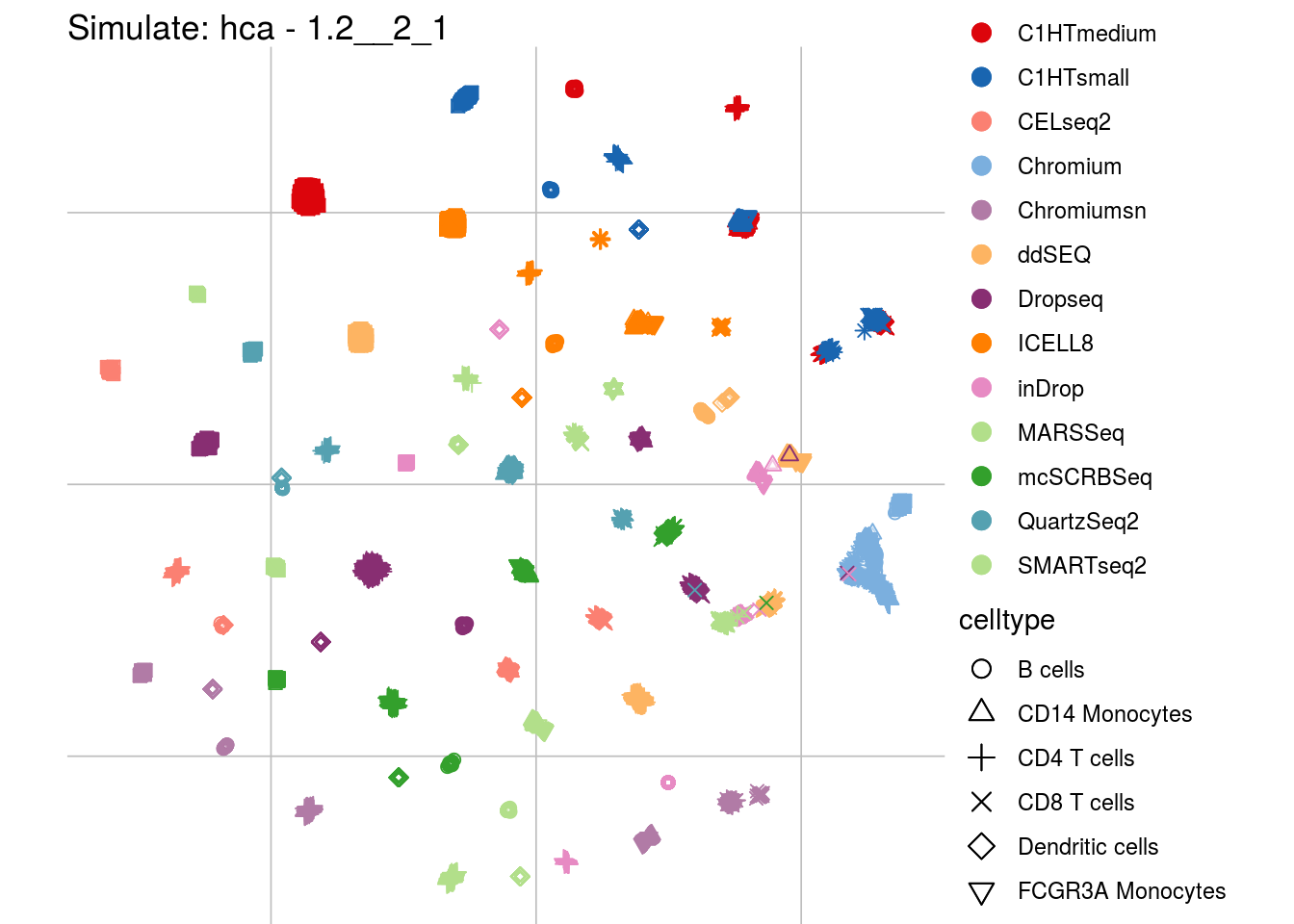
0.8__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.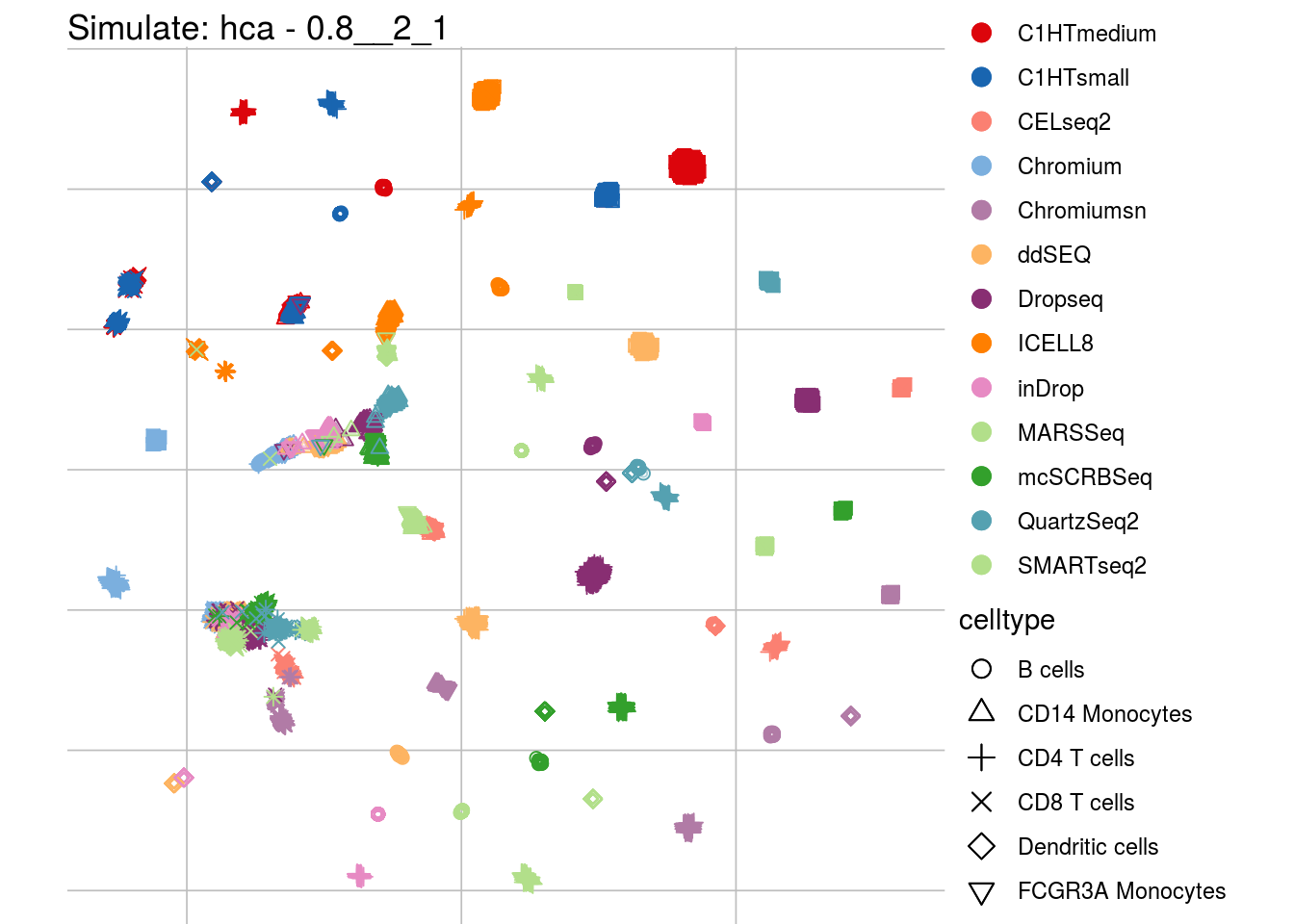
4__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.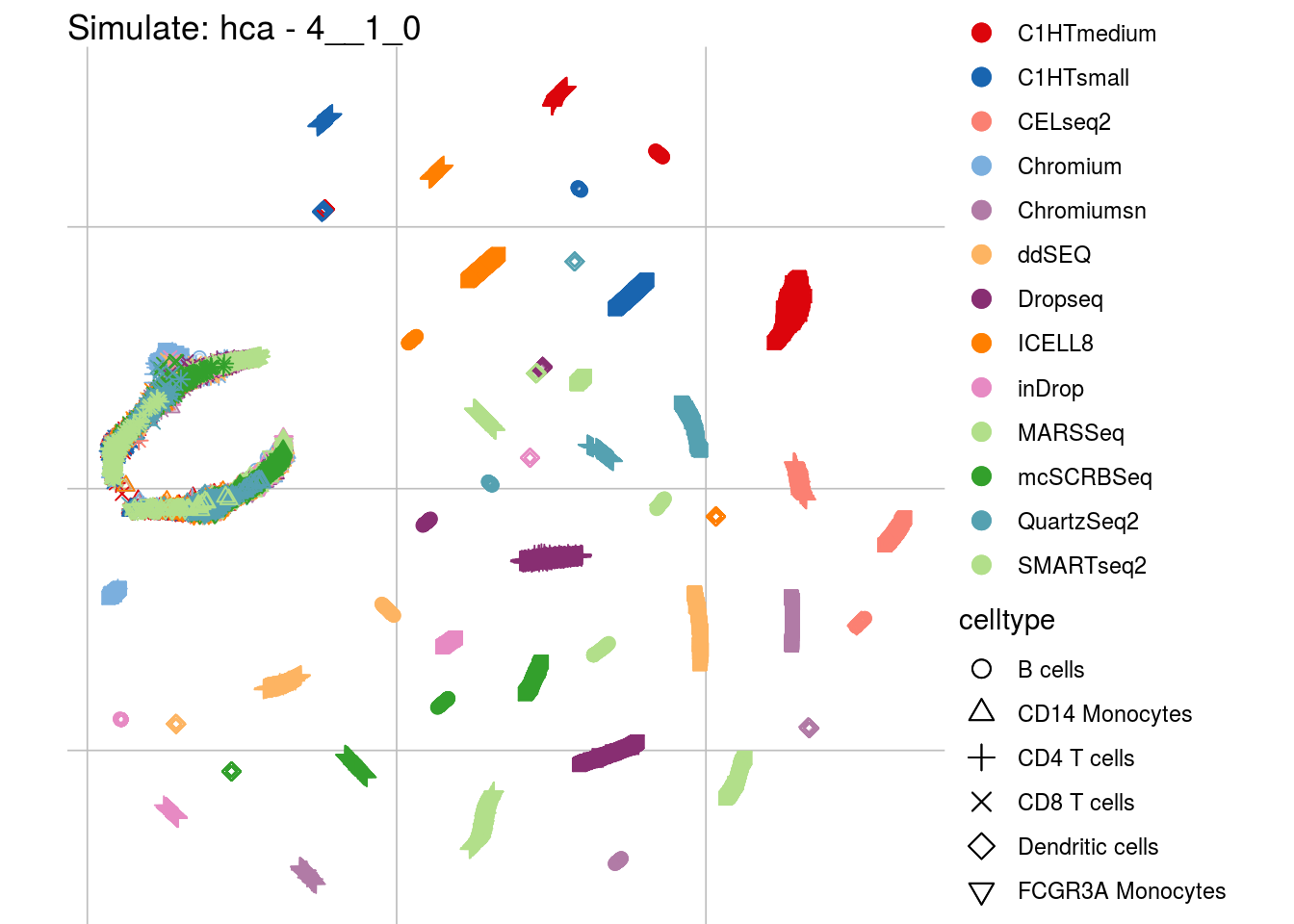
0.3__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.4__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0__0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.8__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.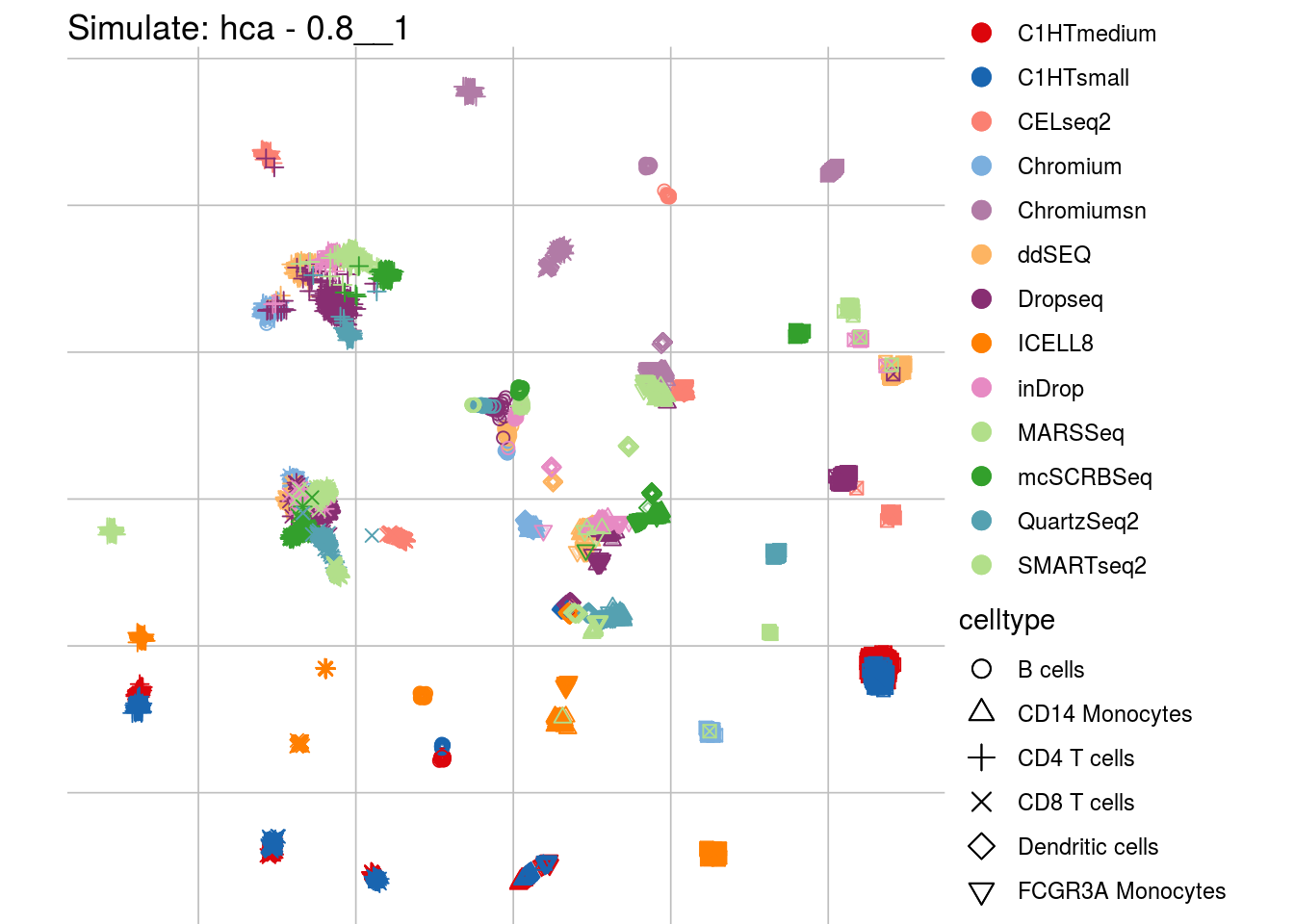
2__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.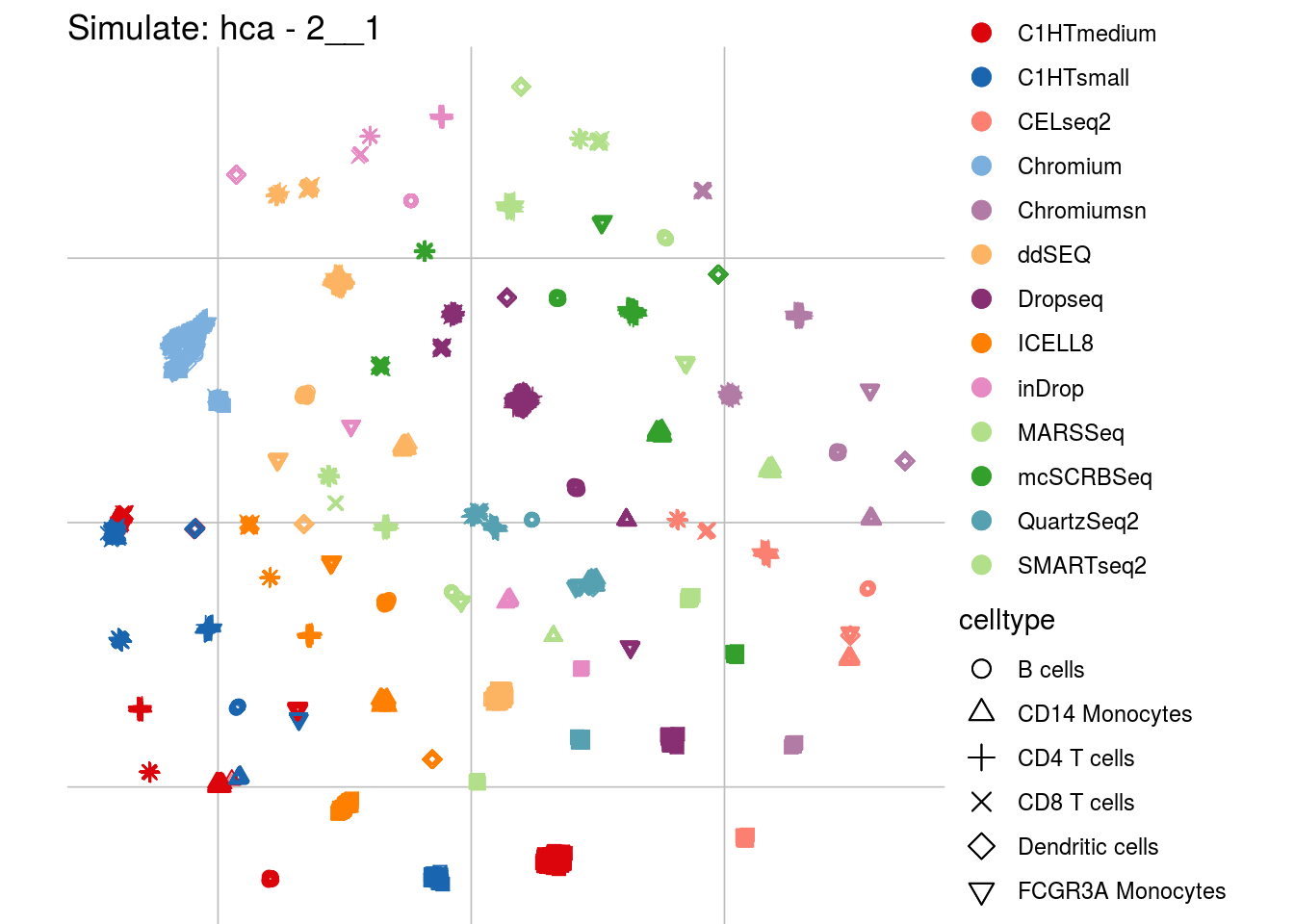
1.5__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.9__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.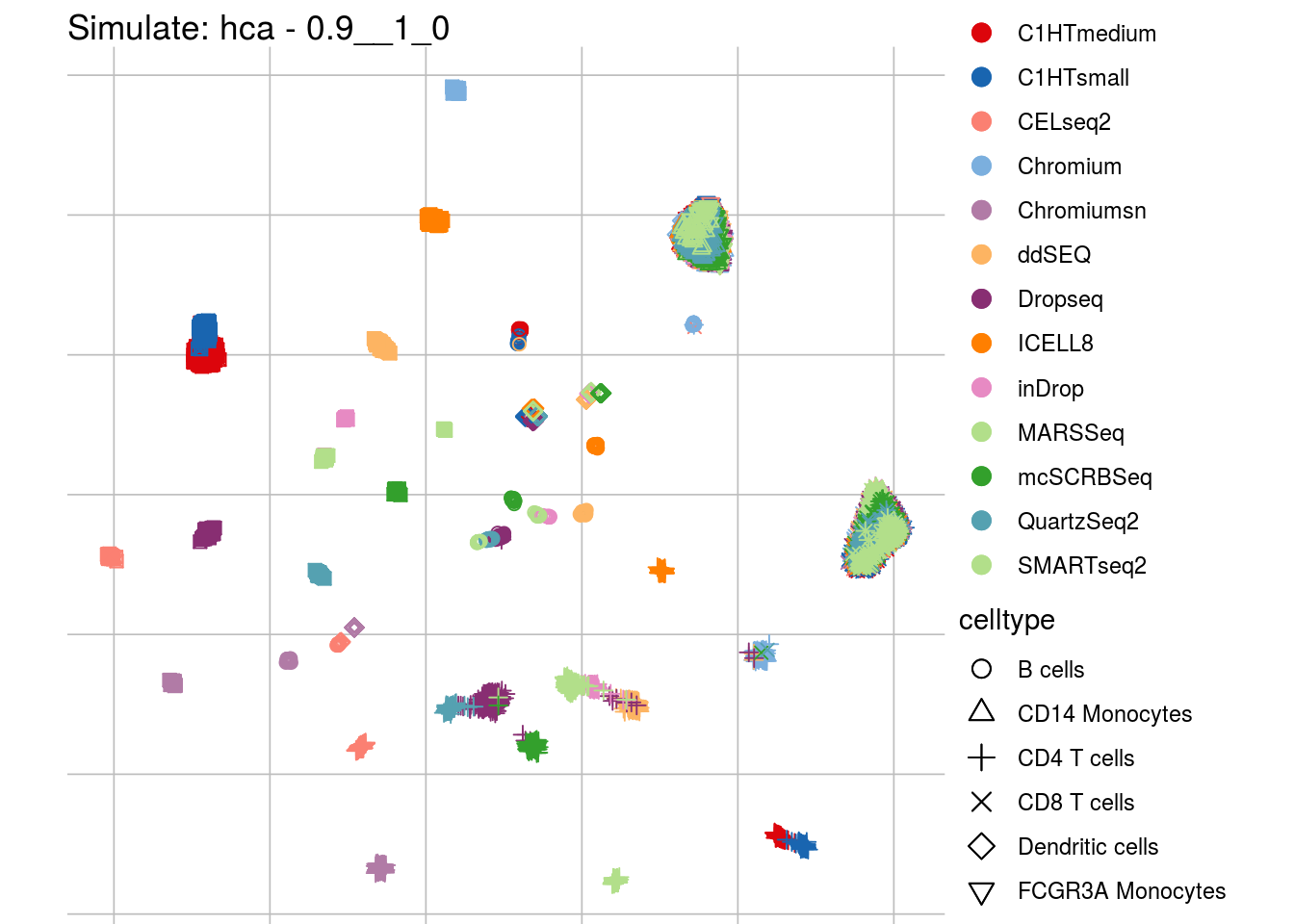
4__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.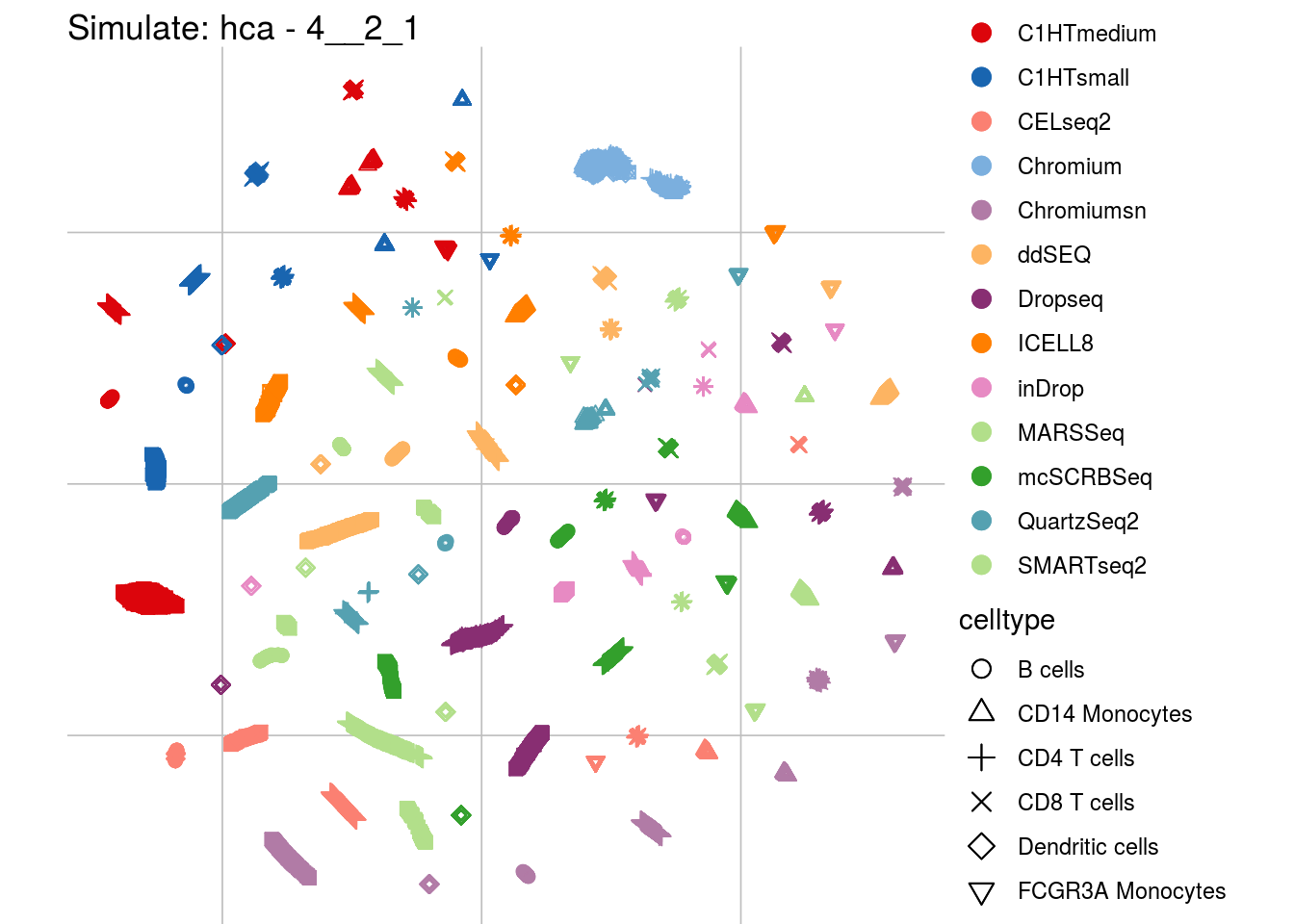
1.2__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.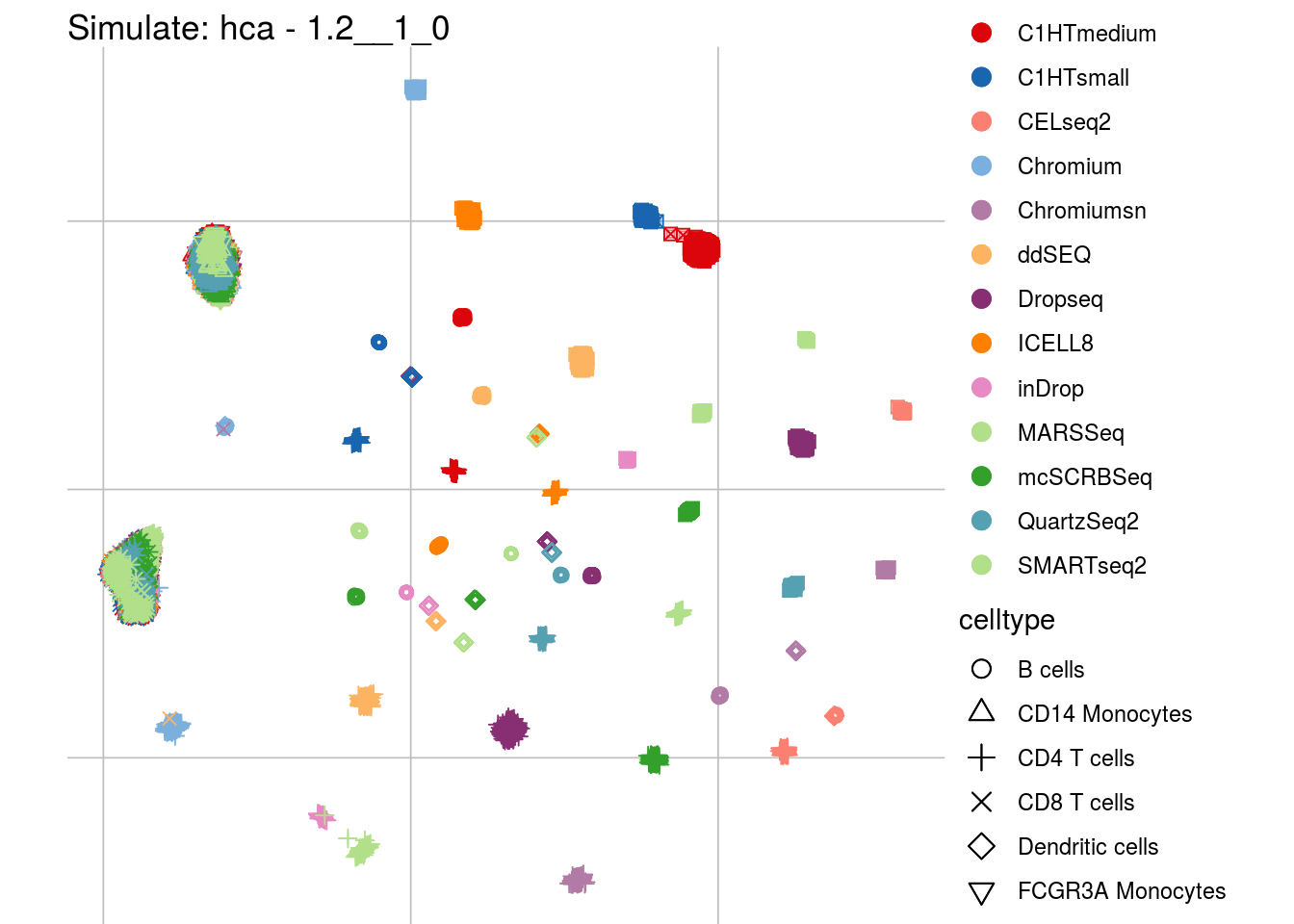
1.5__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.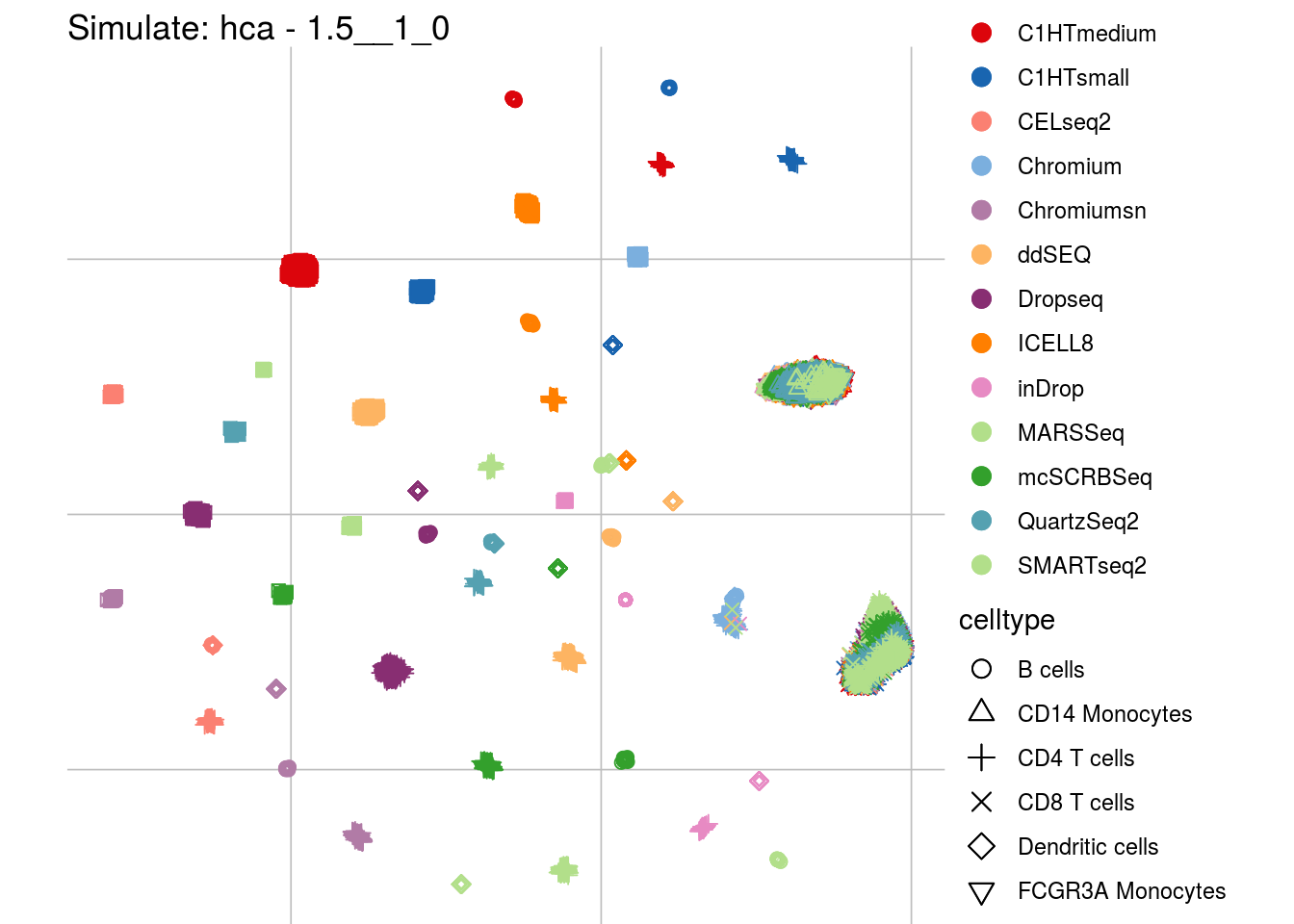
0.6__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.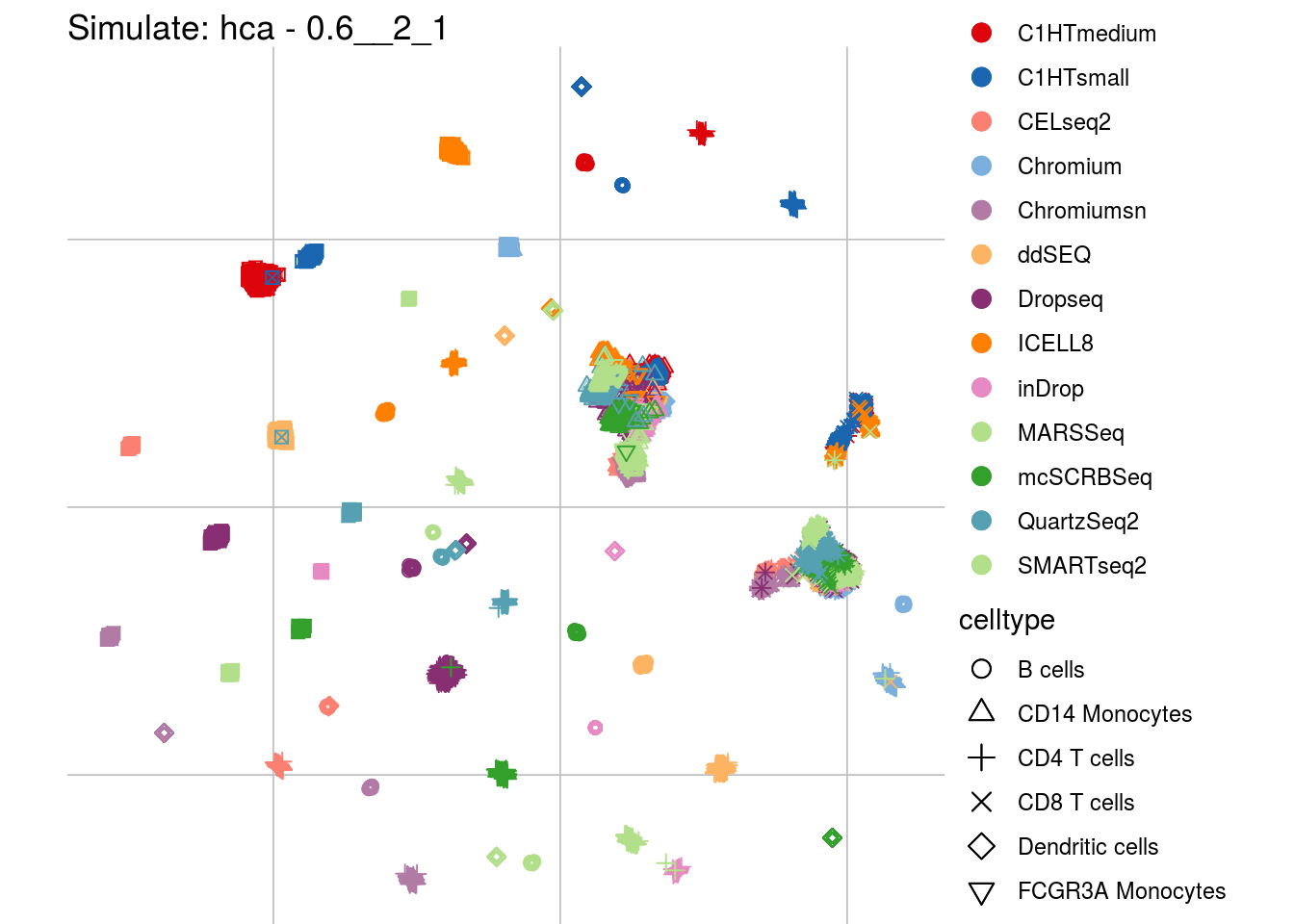
0.4__1_0
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
1.5__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.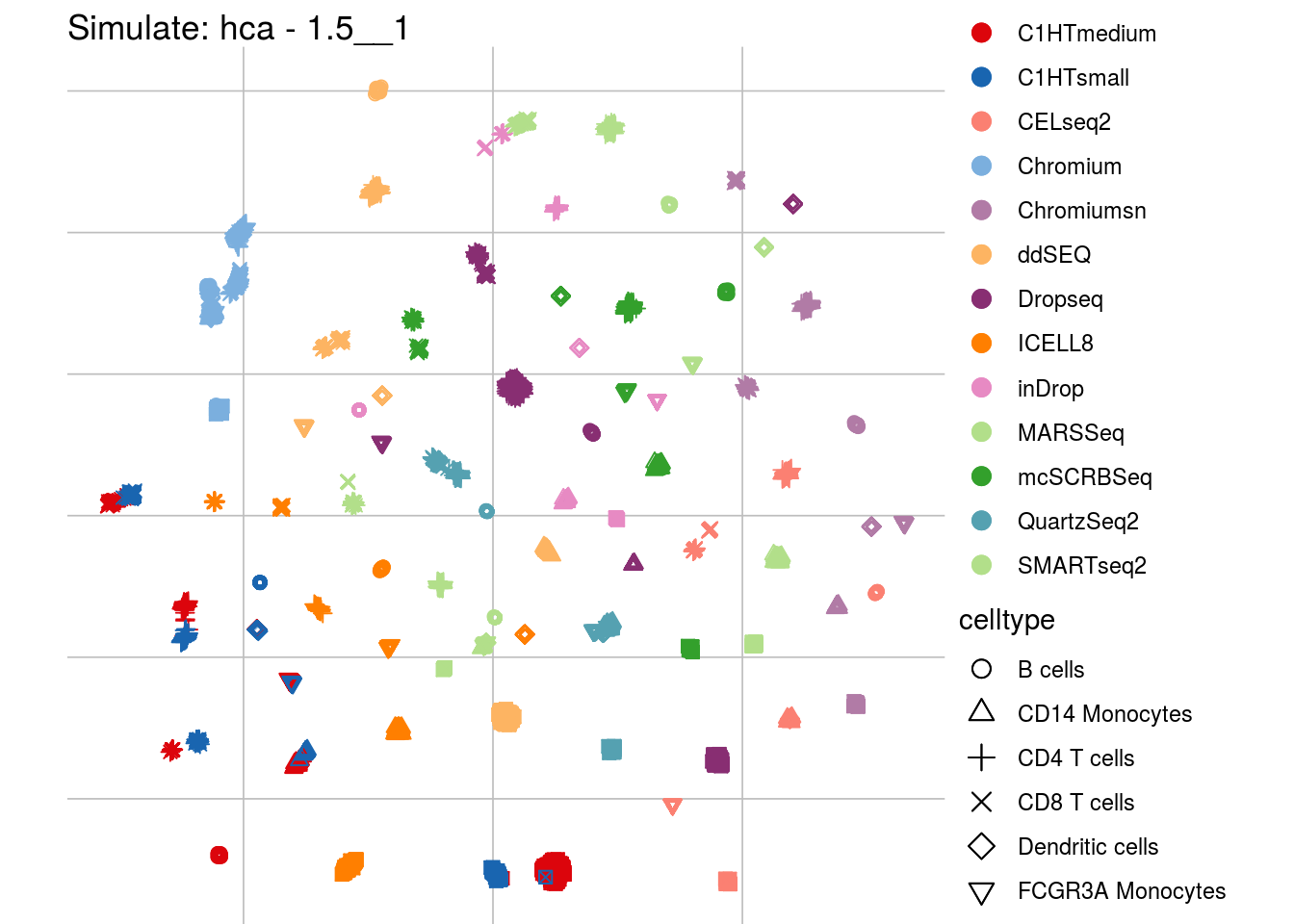
1__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.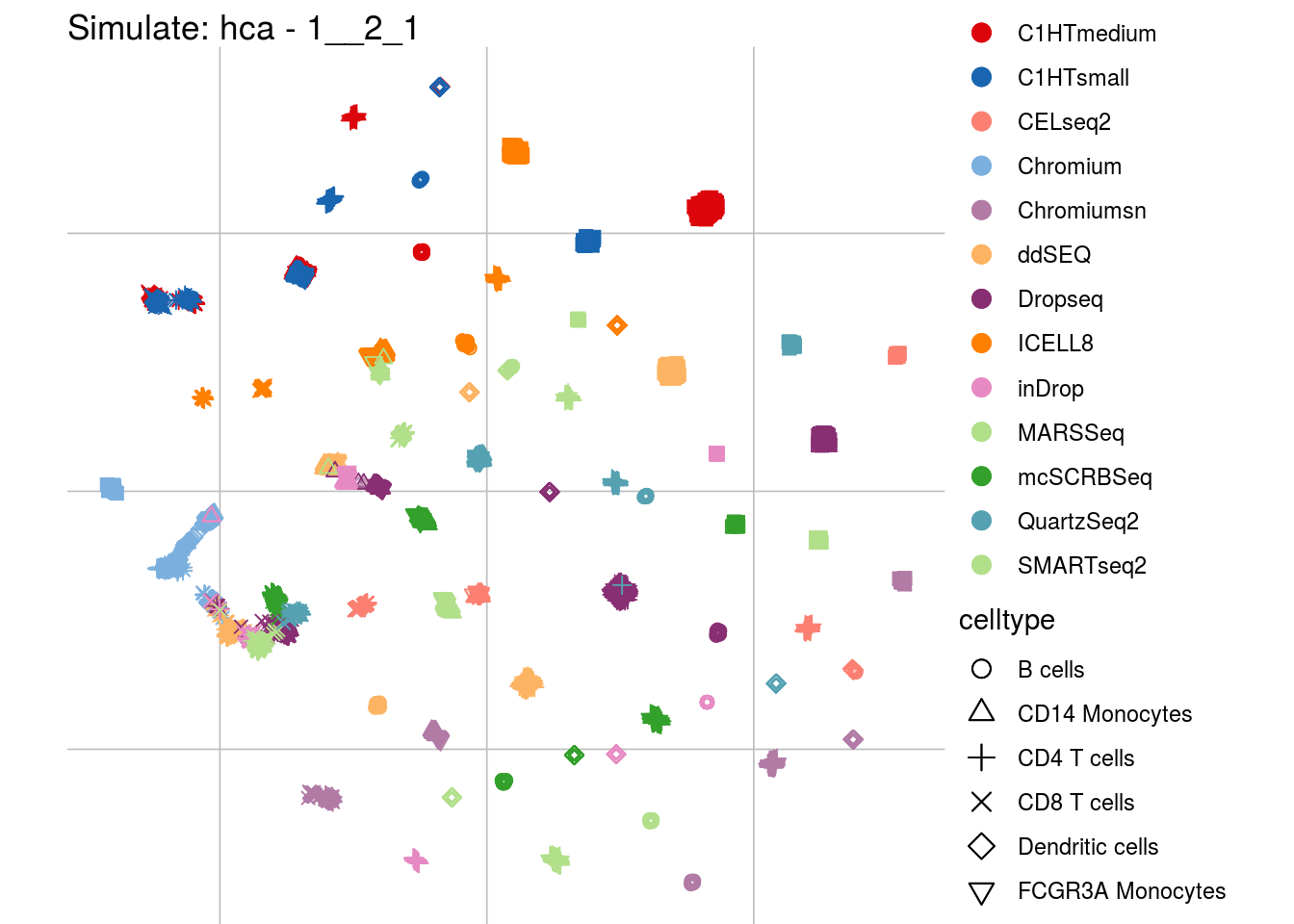
0.7__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.6__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.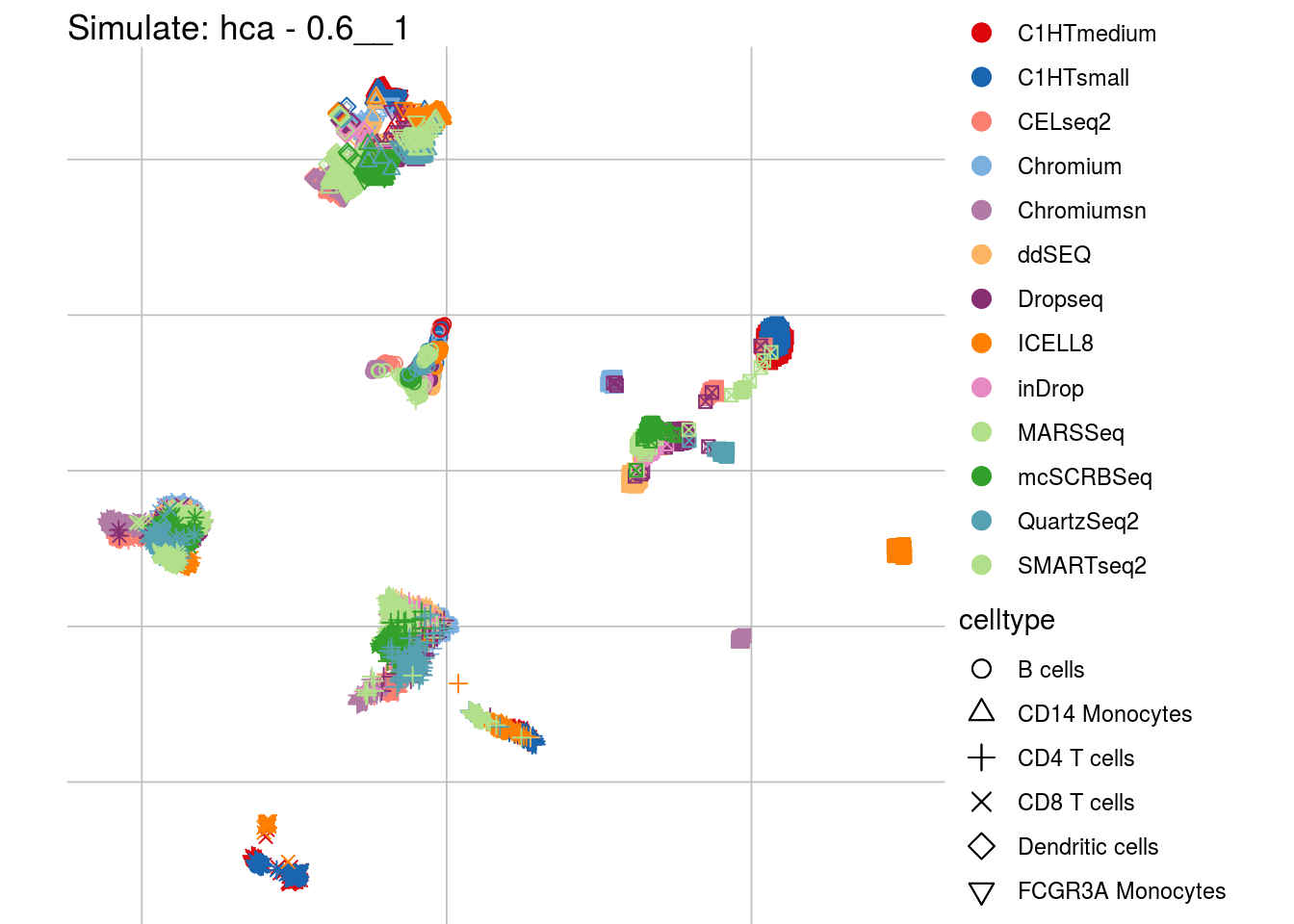
0.9__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.3__2_1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.
0.4__1
## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.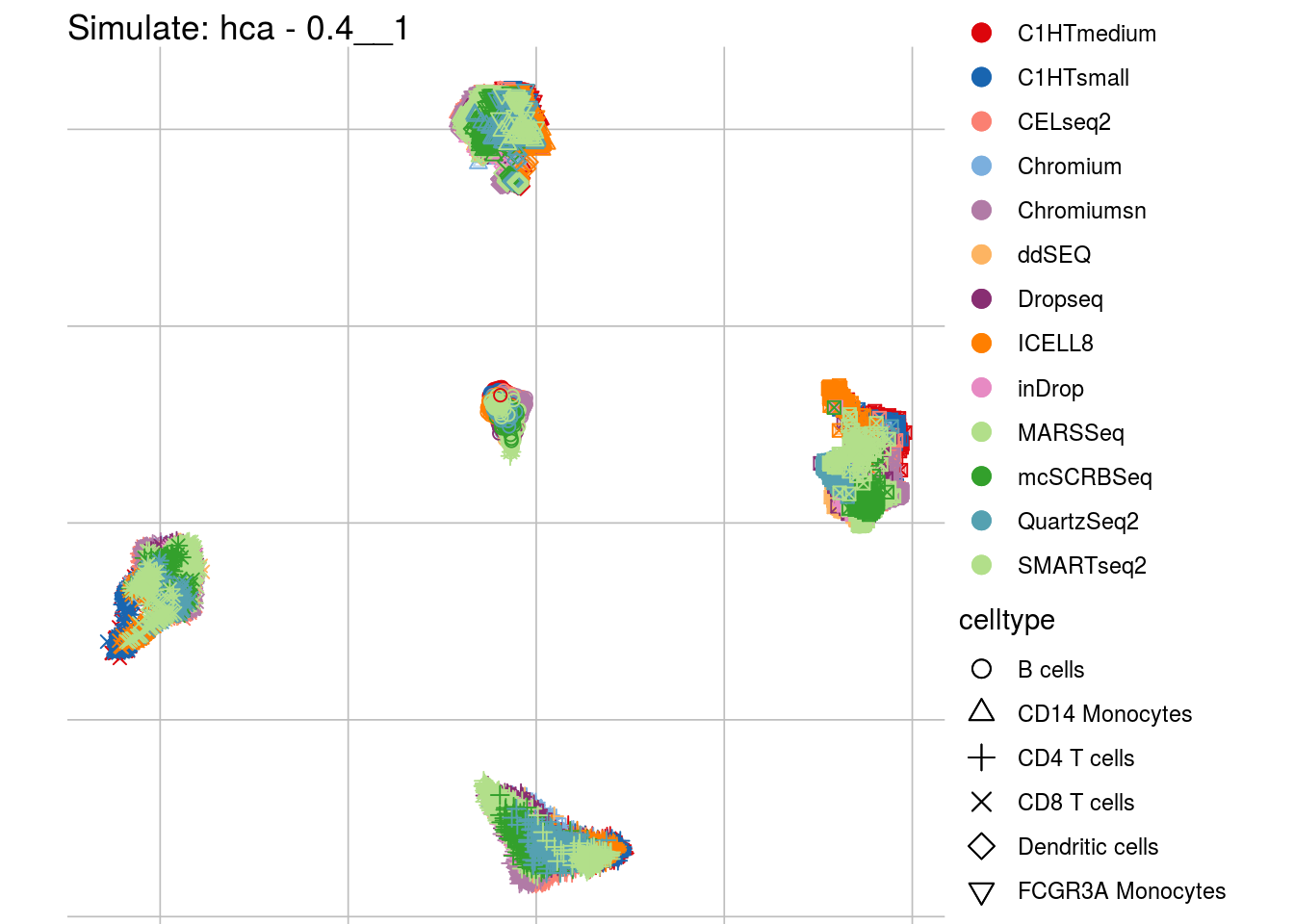
MDS plot
template_mds <- c(
"#### {{nm}}\n",
"```{r mds{{nm}}, echo = FALSE}\n",
"filename <- paste0('out/sim_char/', '{{name}}', '/sim_', '{{name}}', '_', '{{nm}}', '_sce.rds')",
"sim_sce <- readRDS(file = filename)",
"visMDS(sim_sce, title = paste0('Simulate: ', '{{name}}', ' - ', '{{nm}}'), ids = batch)\n",
"```\n",
"\n"
)
plots_mds <- lapply(sim_list[[1]][[1]],
function(nm, name = dataset_name) knitr::knit_expand(text = template_mds)
)1__1_0

2__1_0
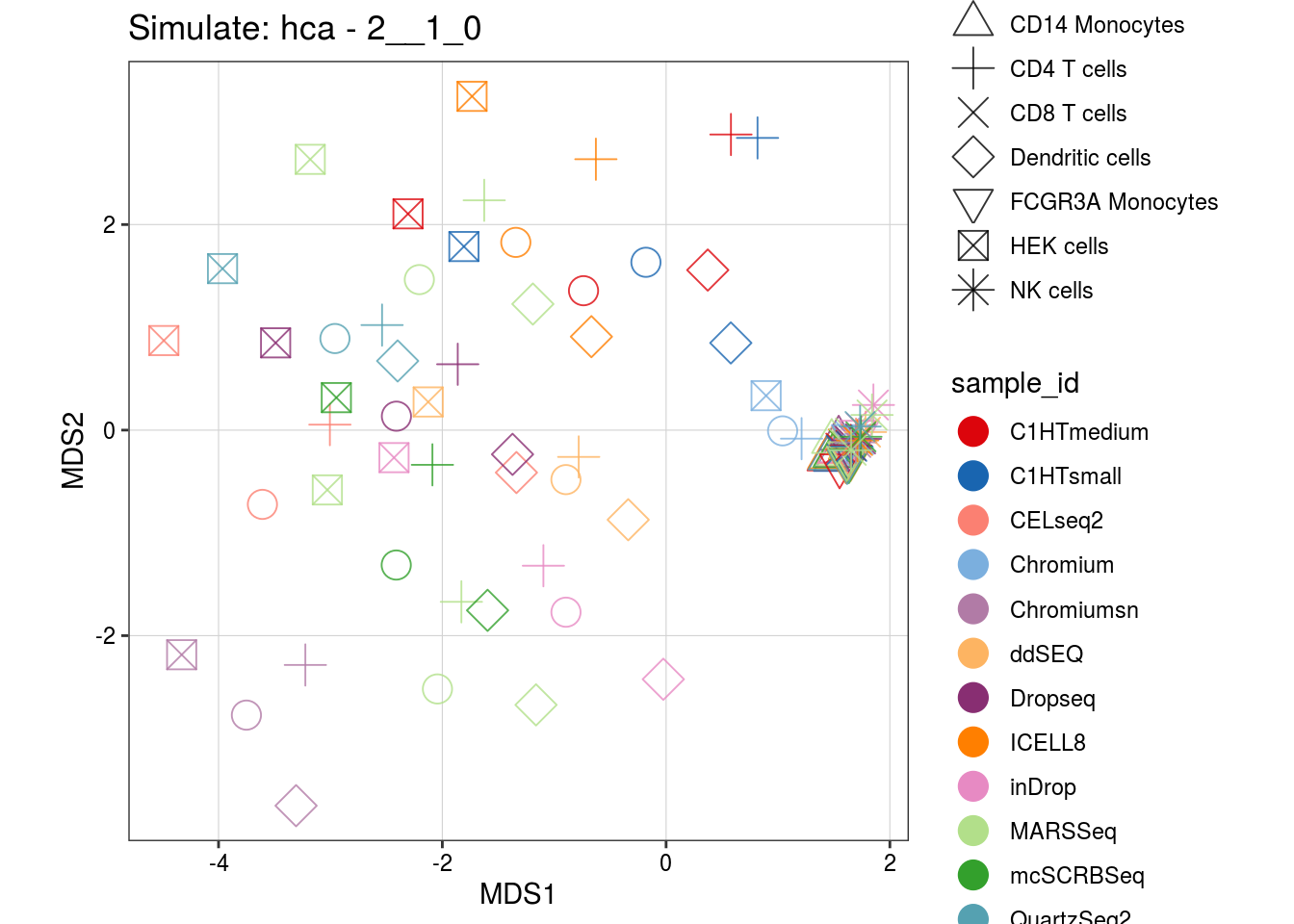
0.9__2_1

0.5__1
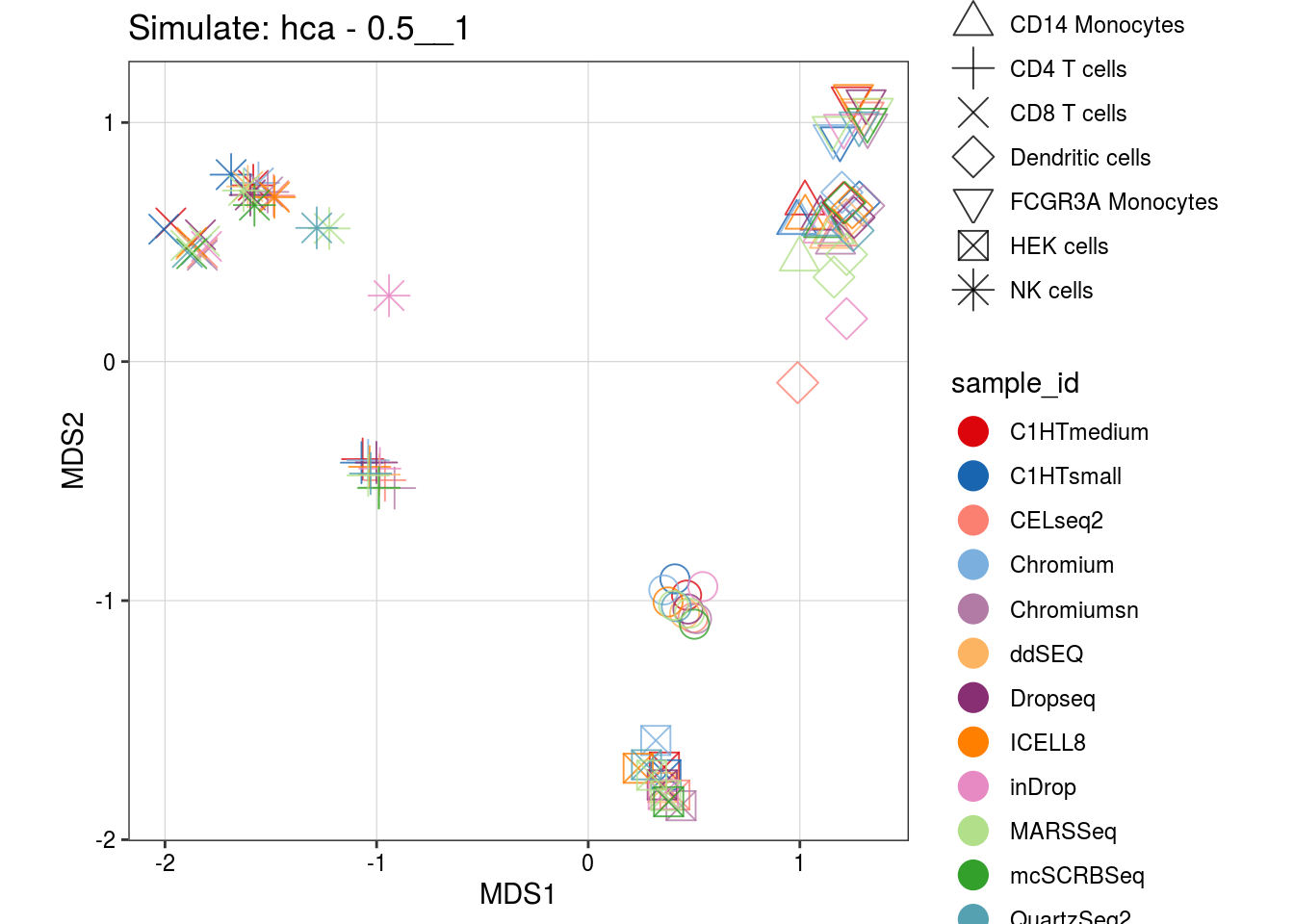
4__1

0.7__1_0
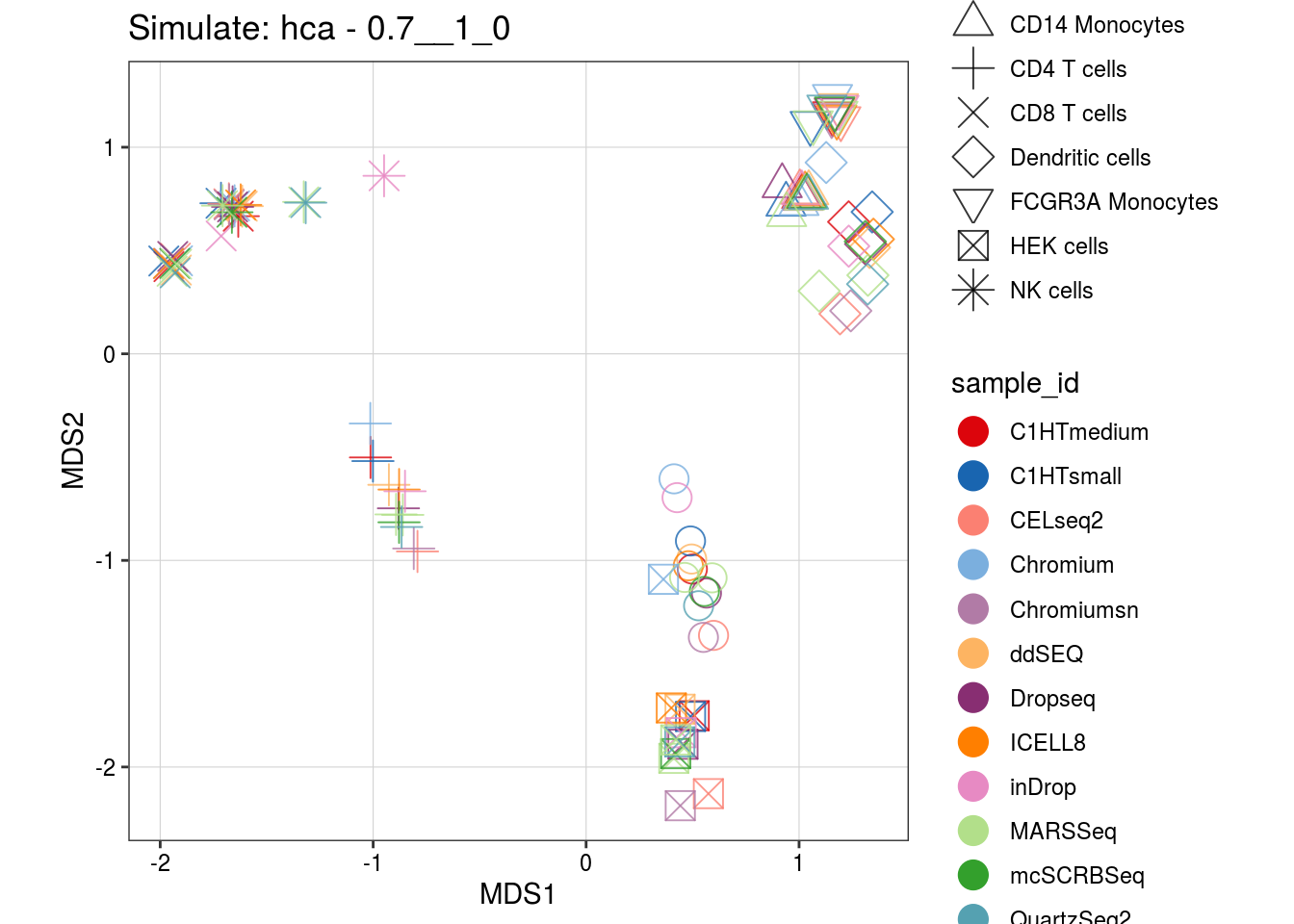
2__2_1

0.7__2_1
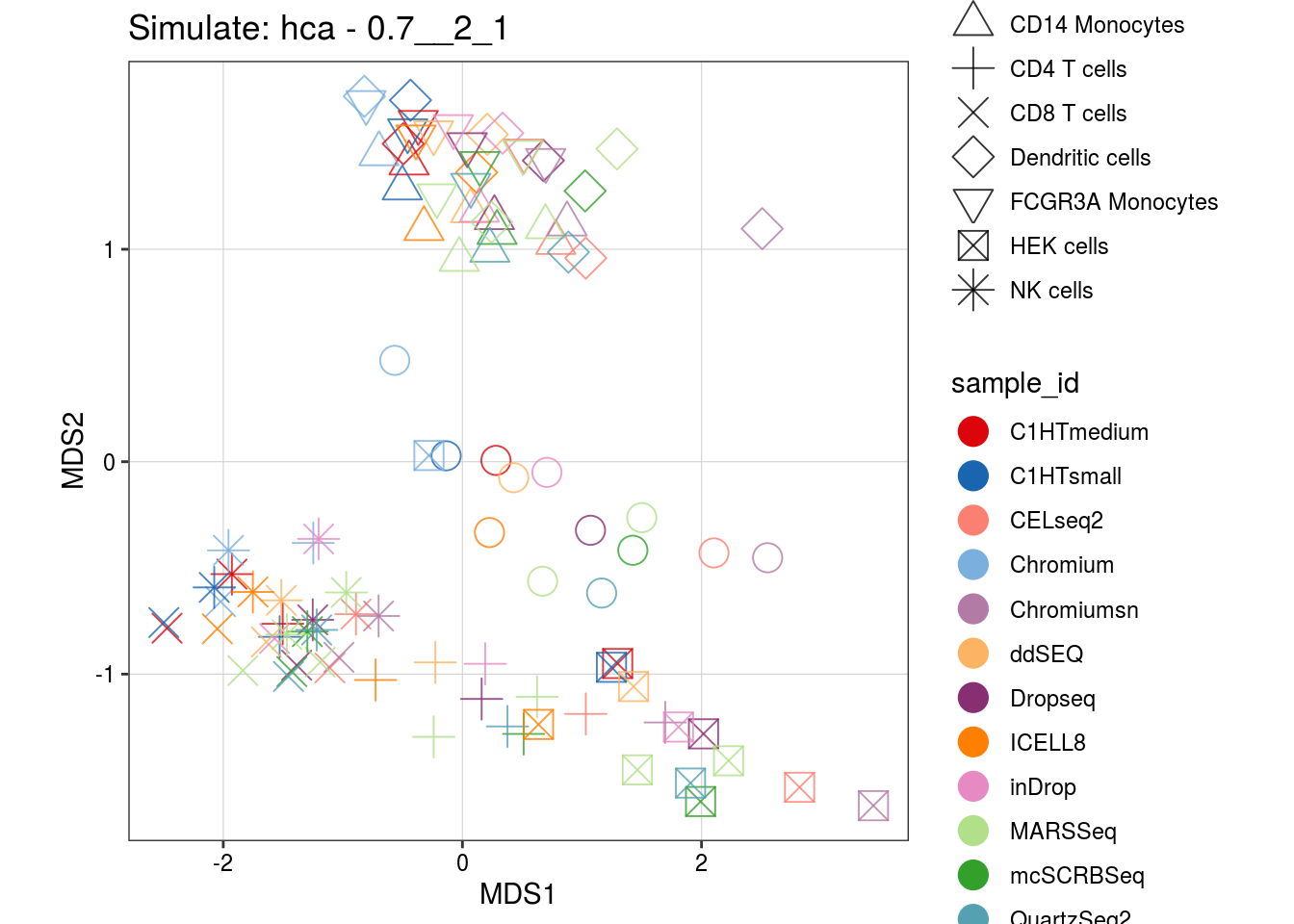
0.5__2_1

0.5__1_0
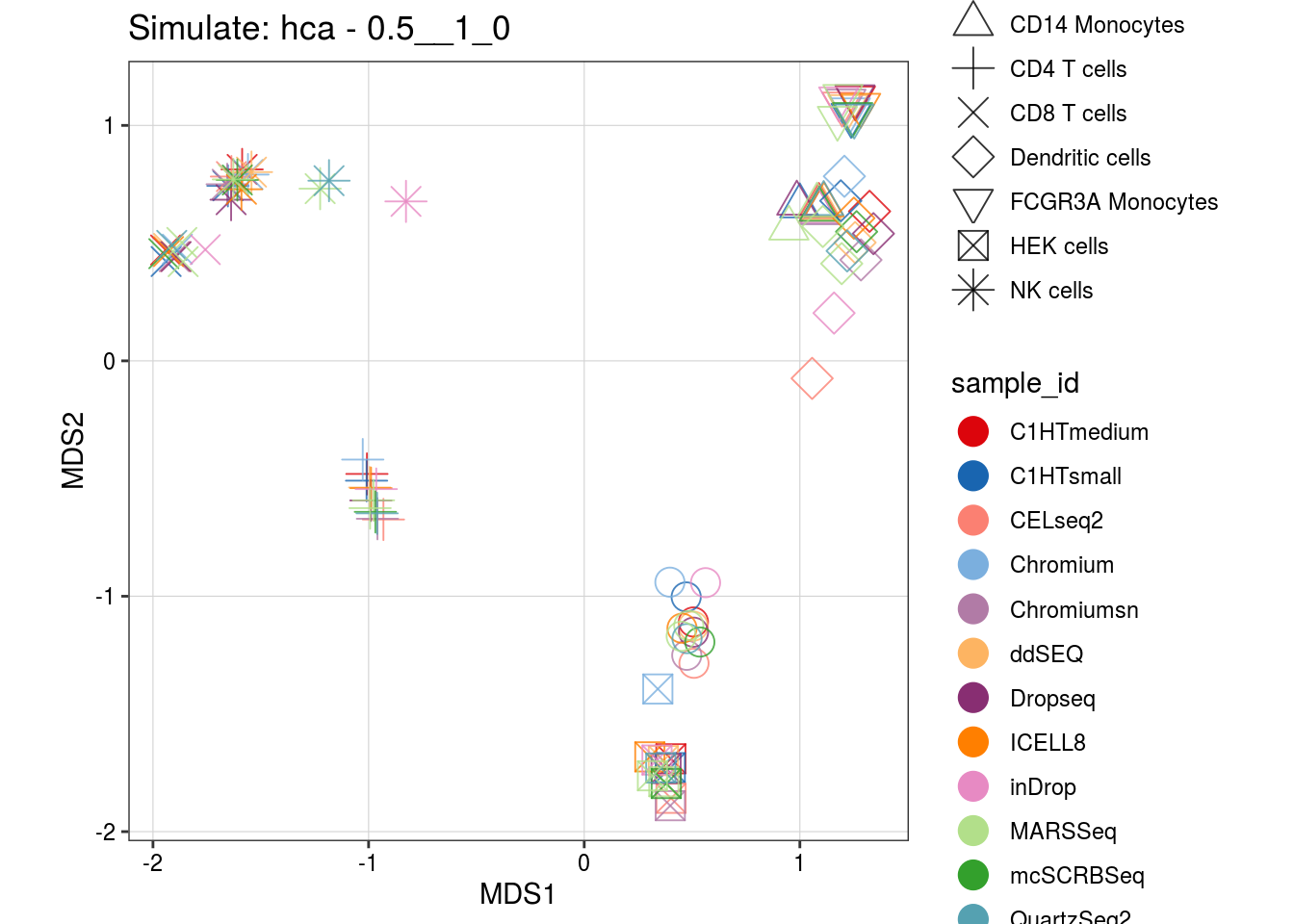
0.3__1

1.2__1
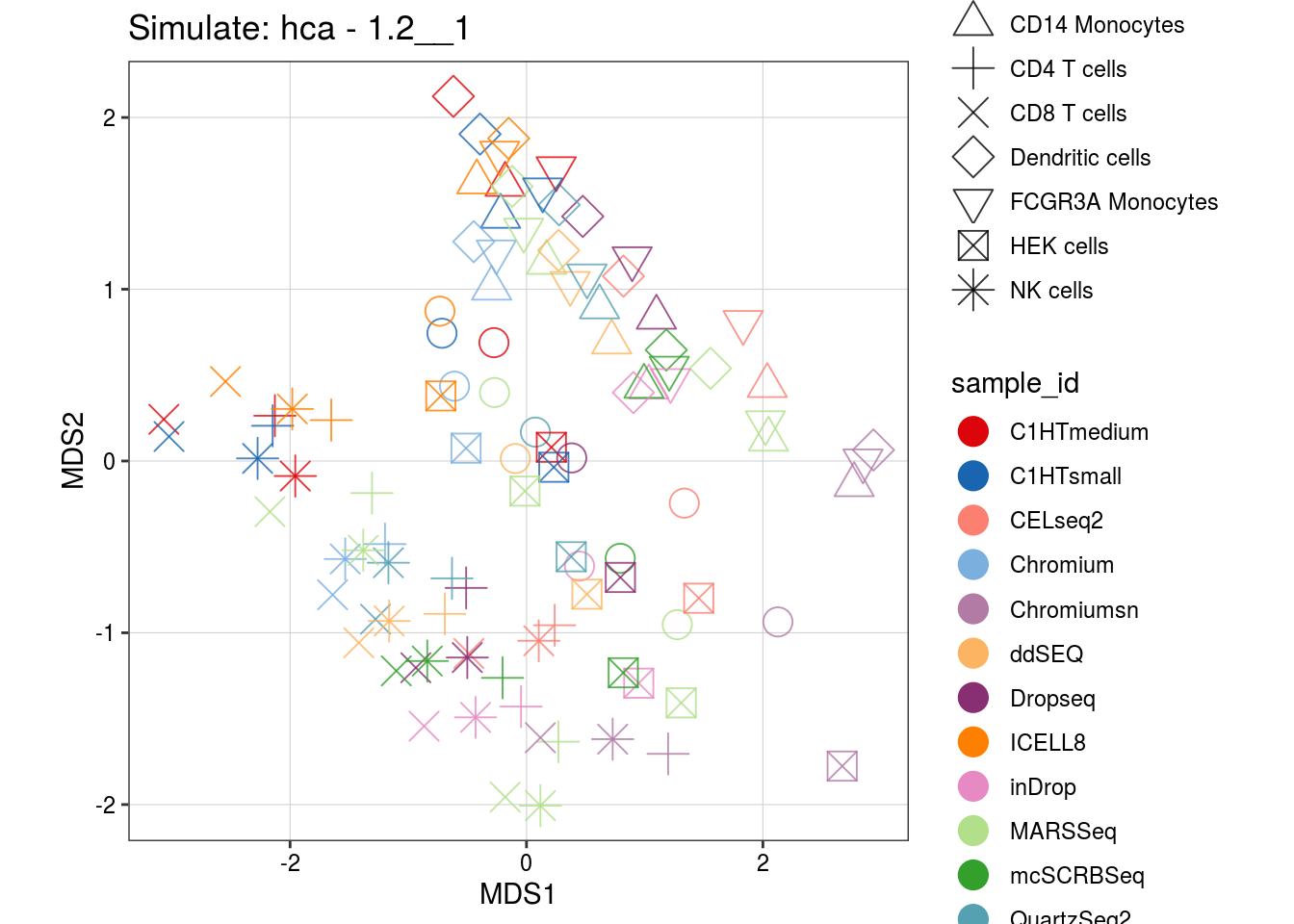
0.8__1_0
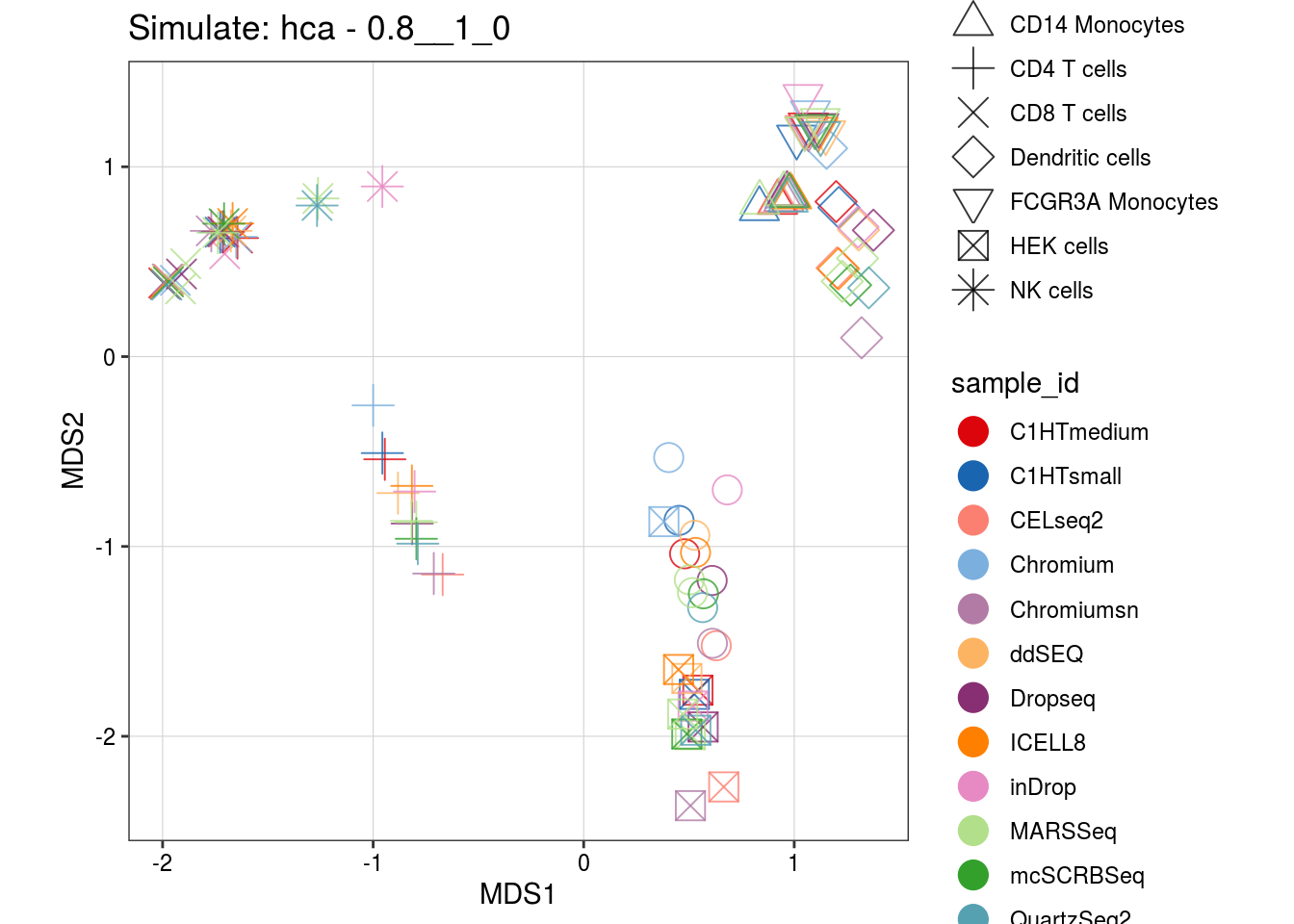
0.6__1_0

1__1

1.2__2_1
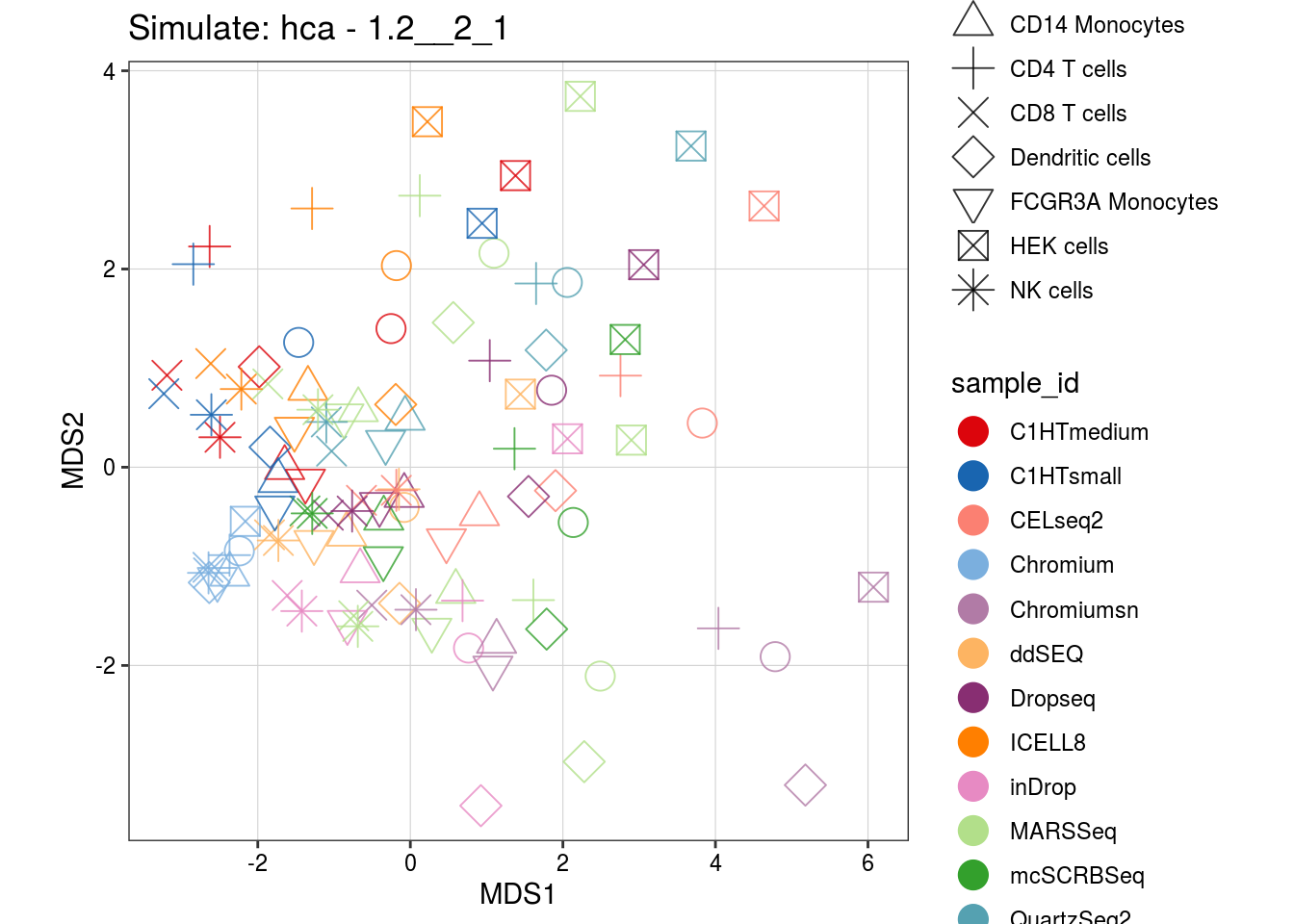
0.8__2_1
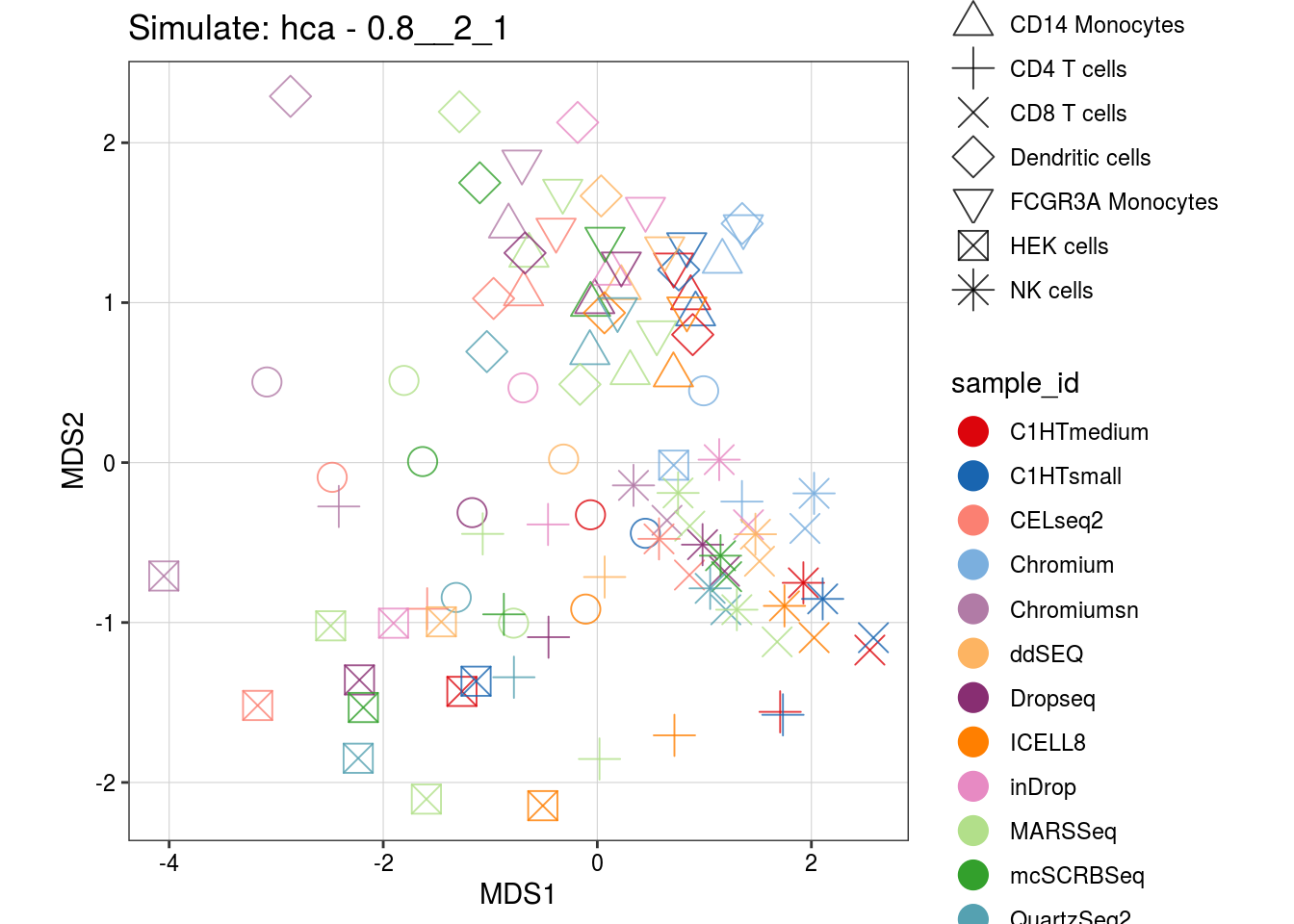
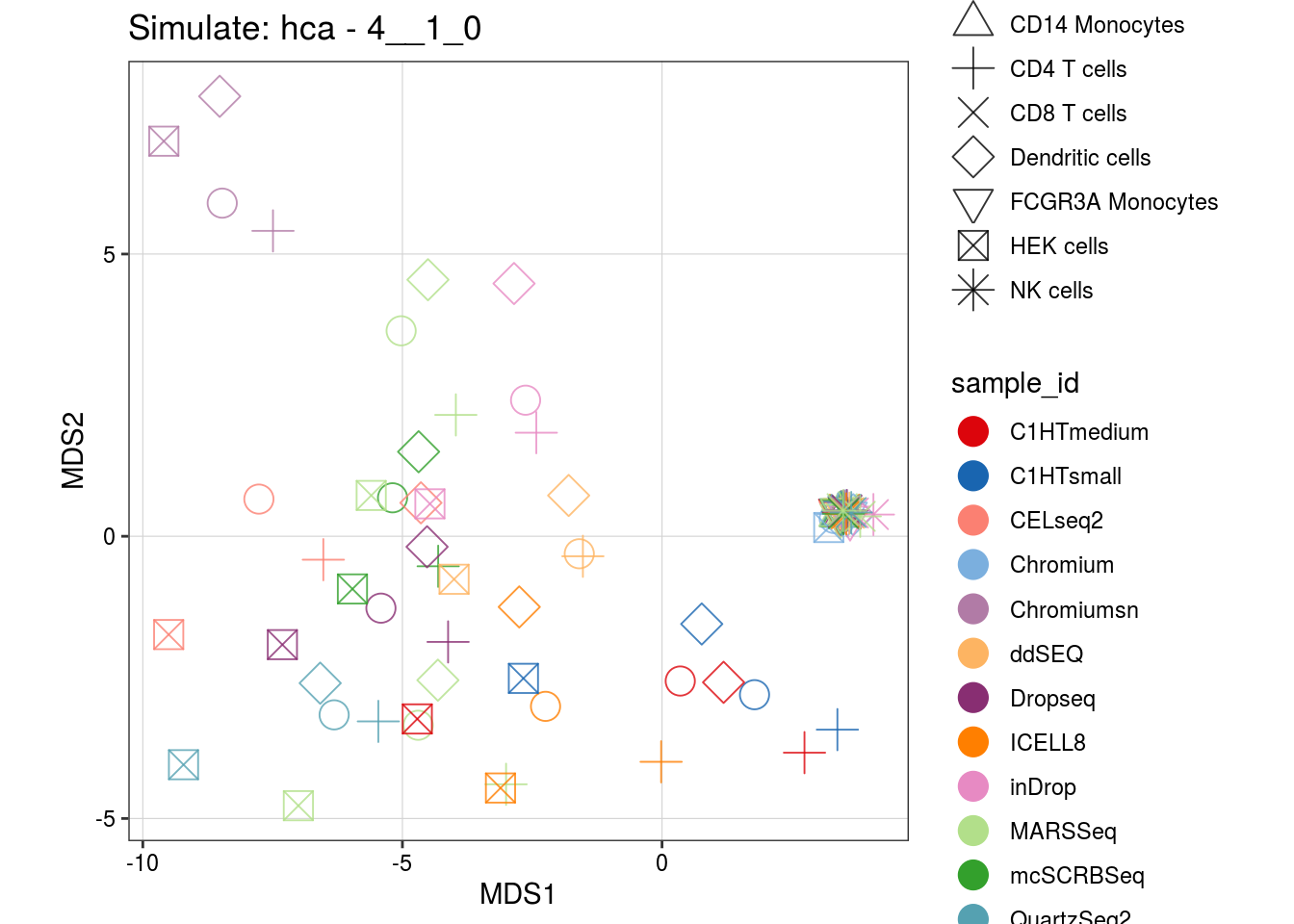
4__1_0
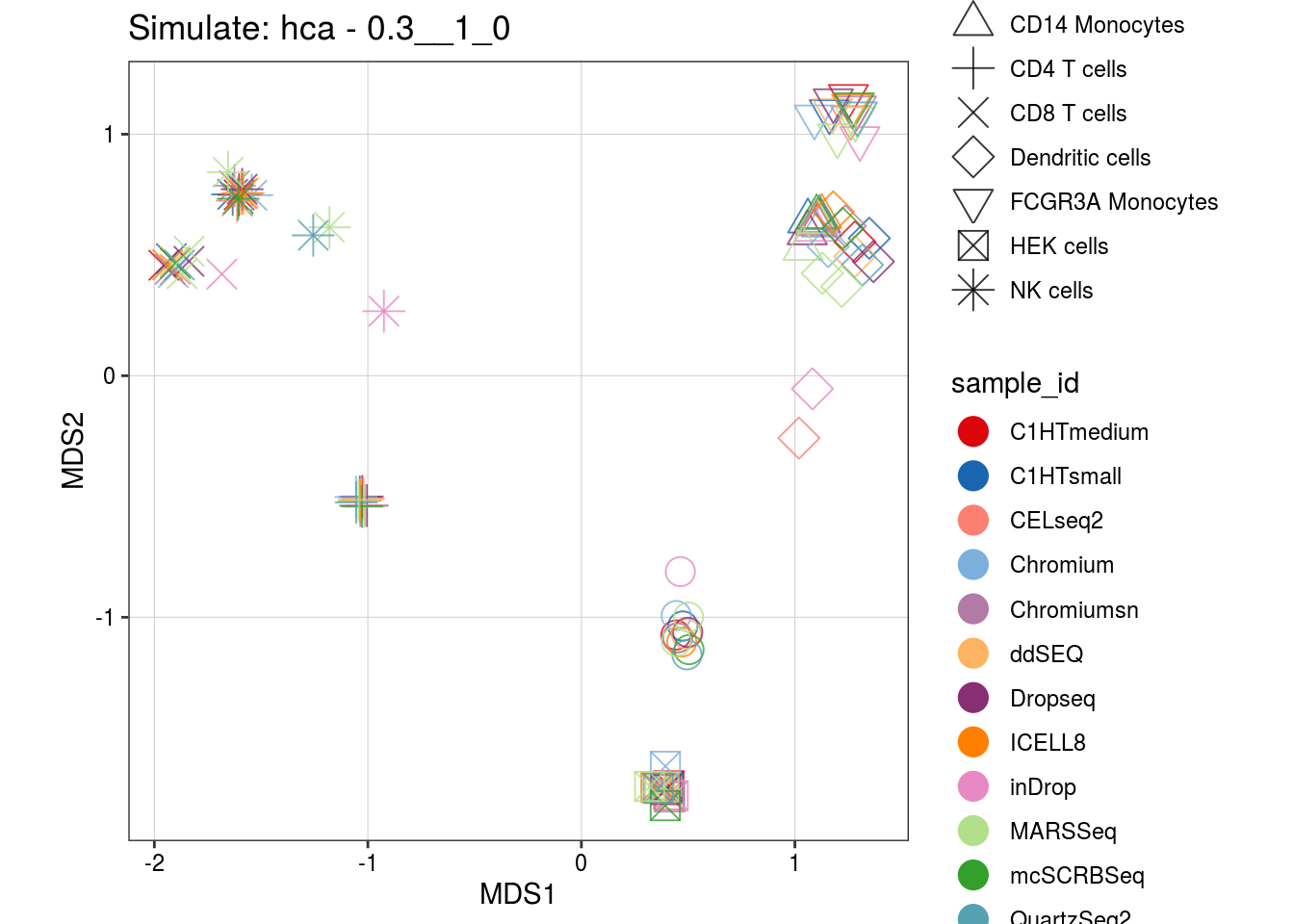
0.3__1_0
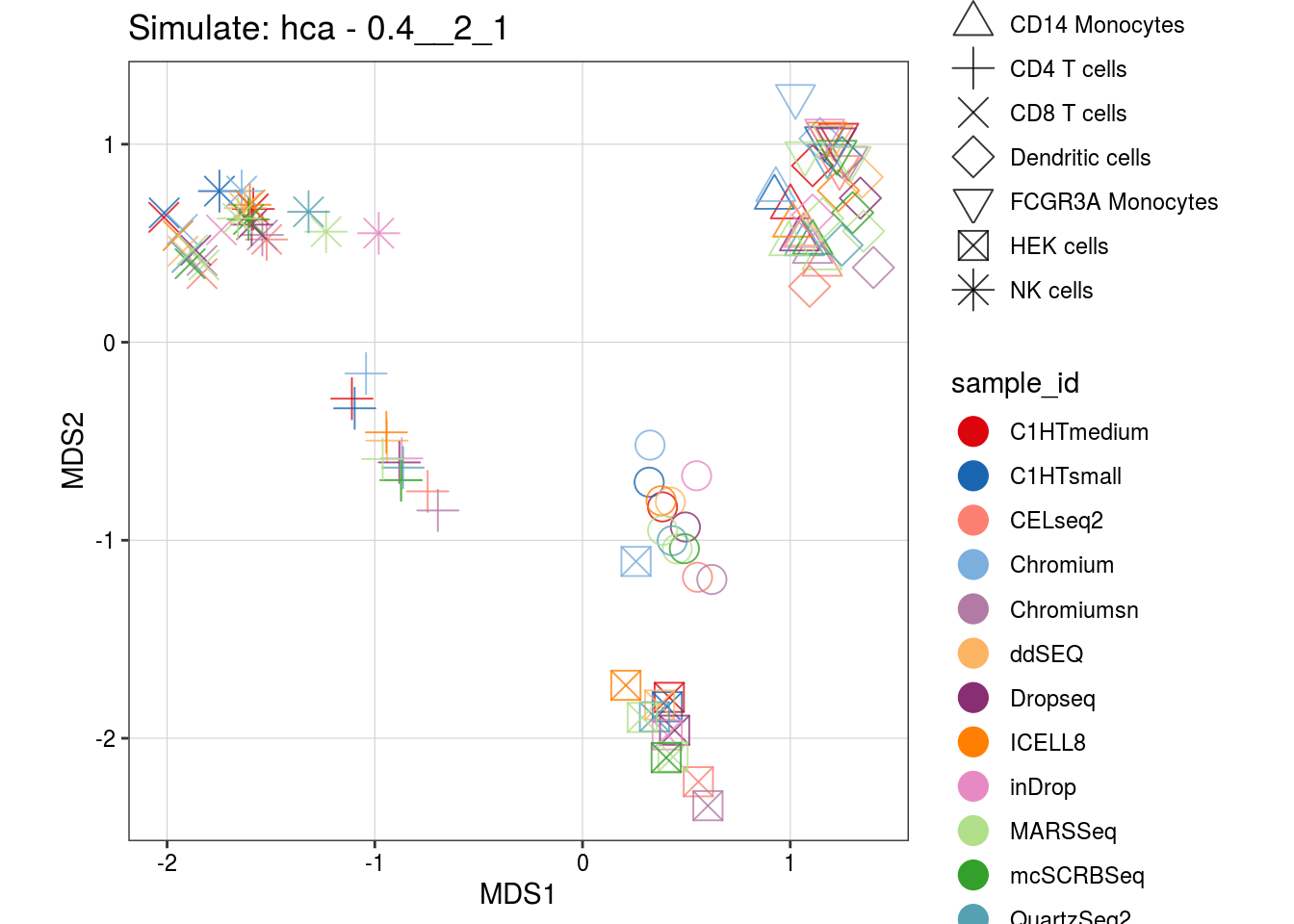
0.4__2_1
0__0

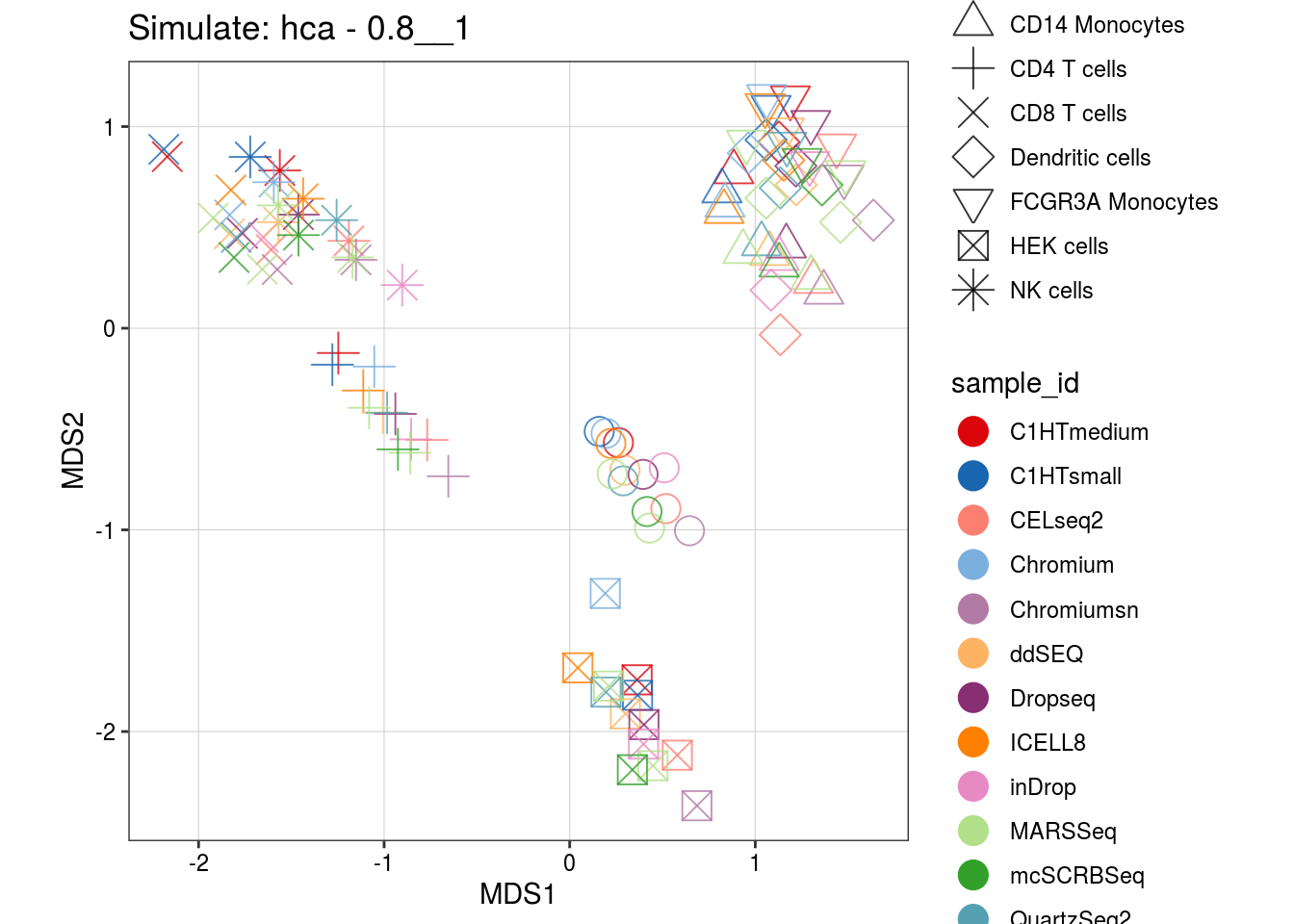
0.8__1
2__1
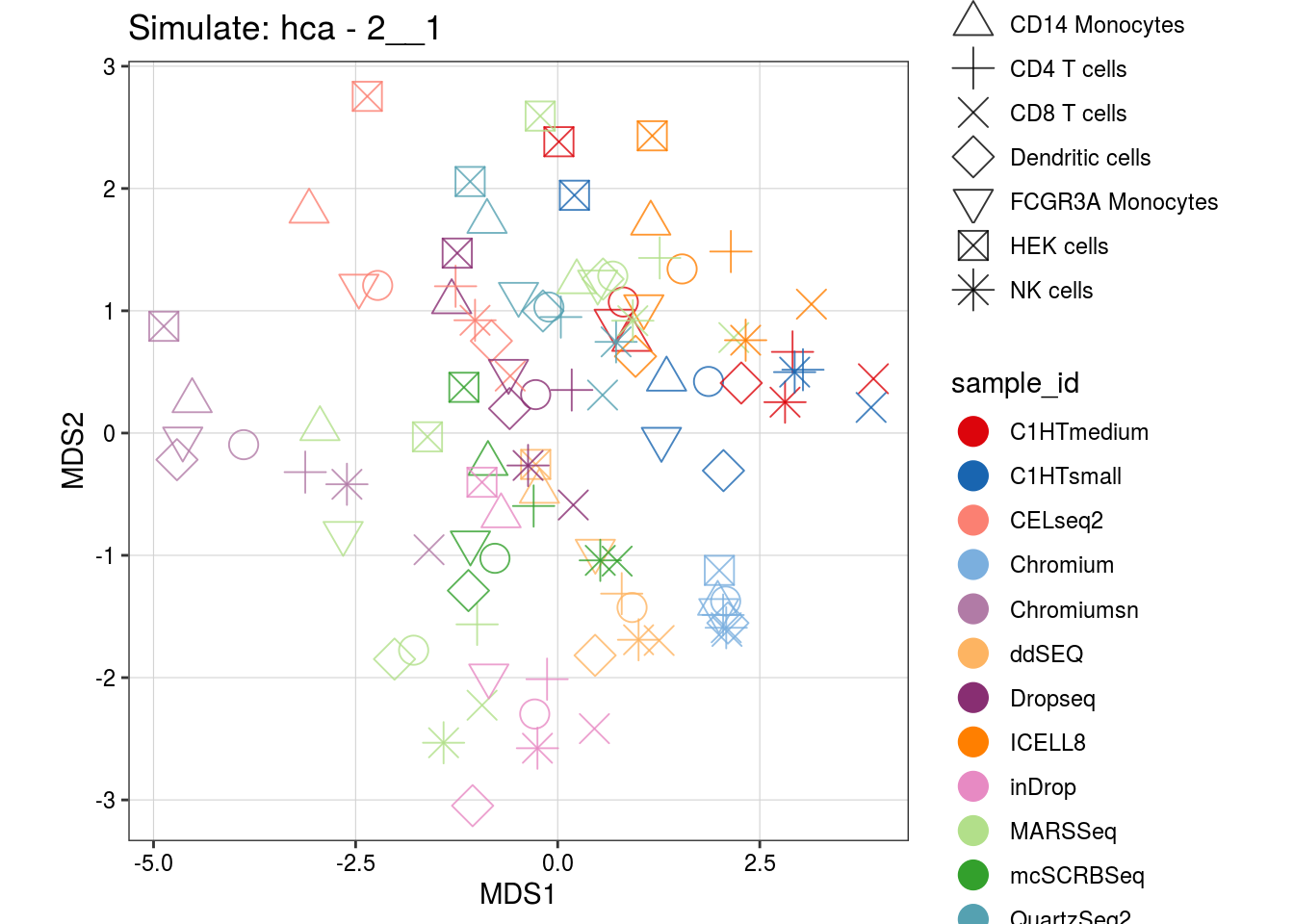
1.5__2_1
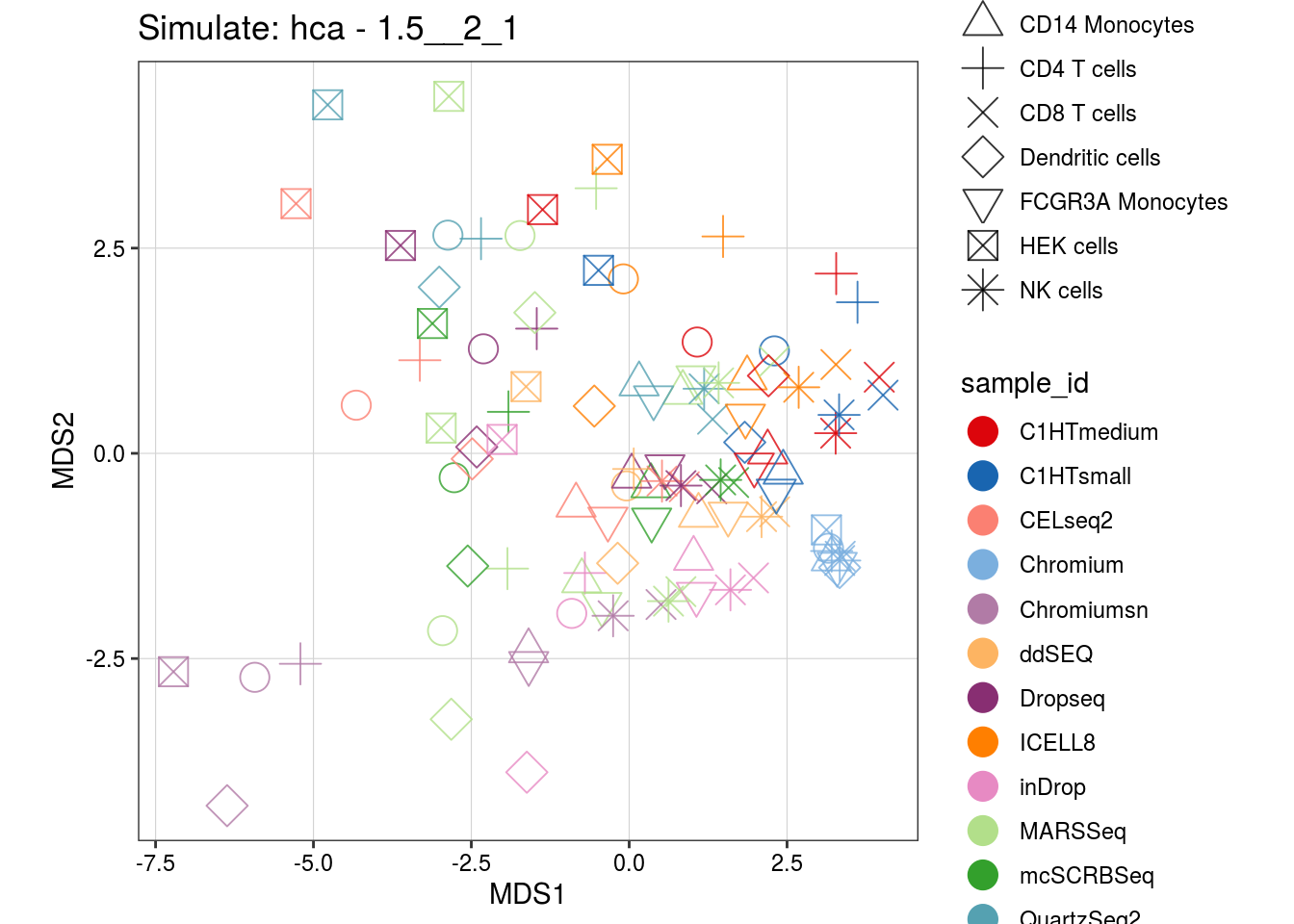
0.9__1_0
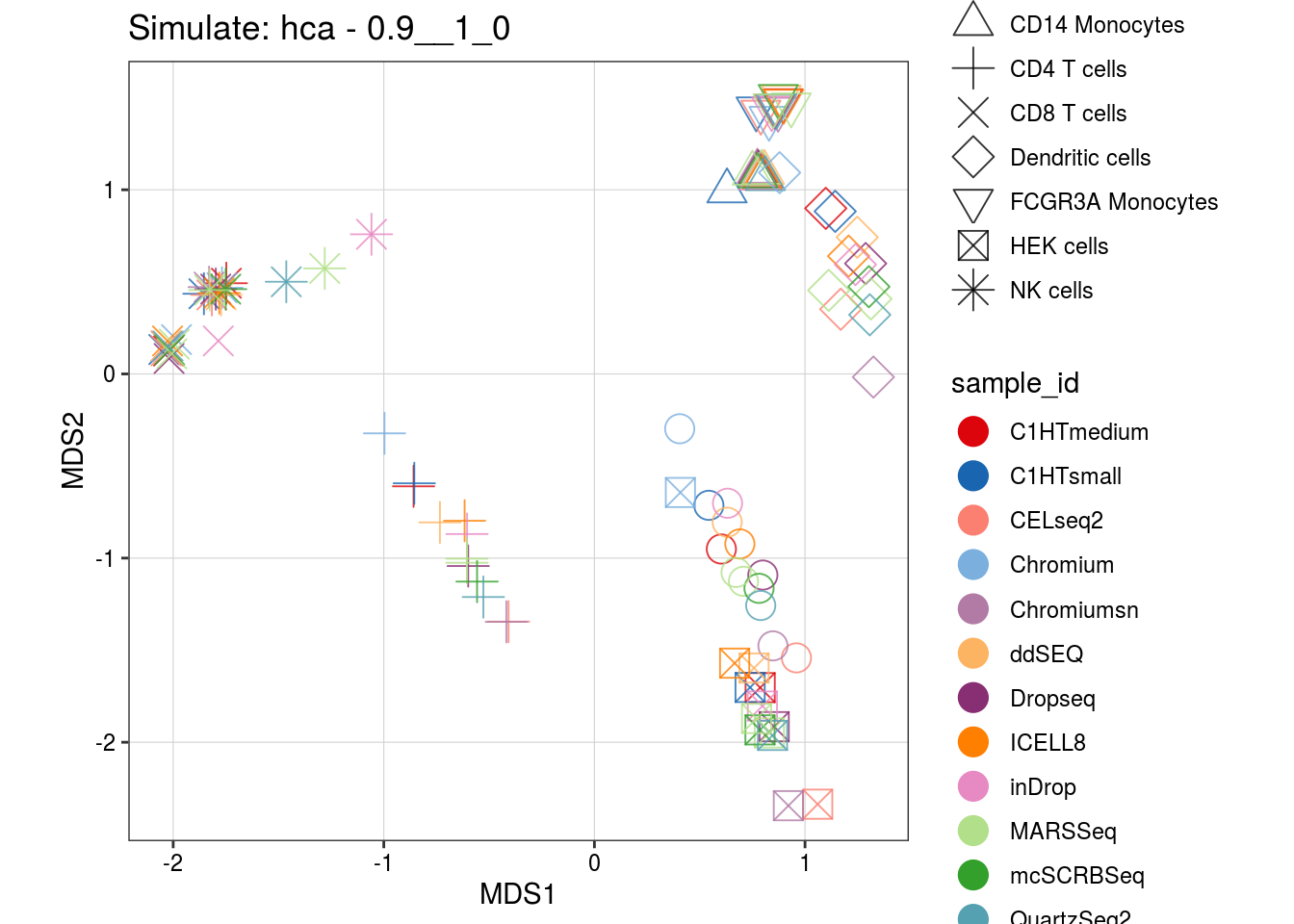
4__2_1
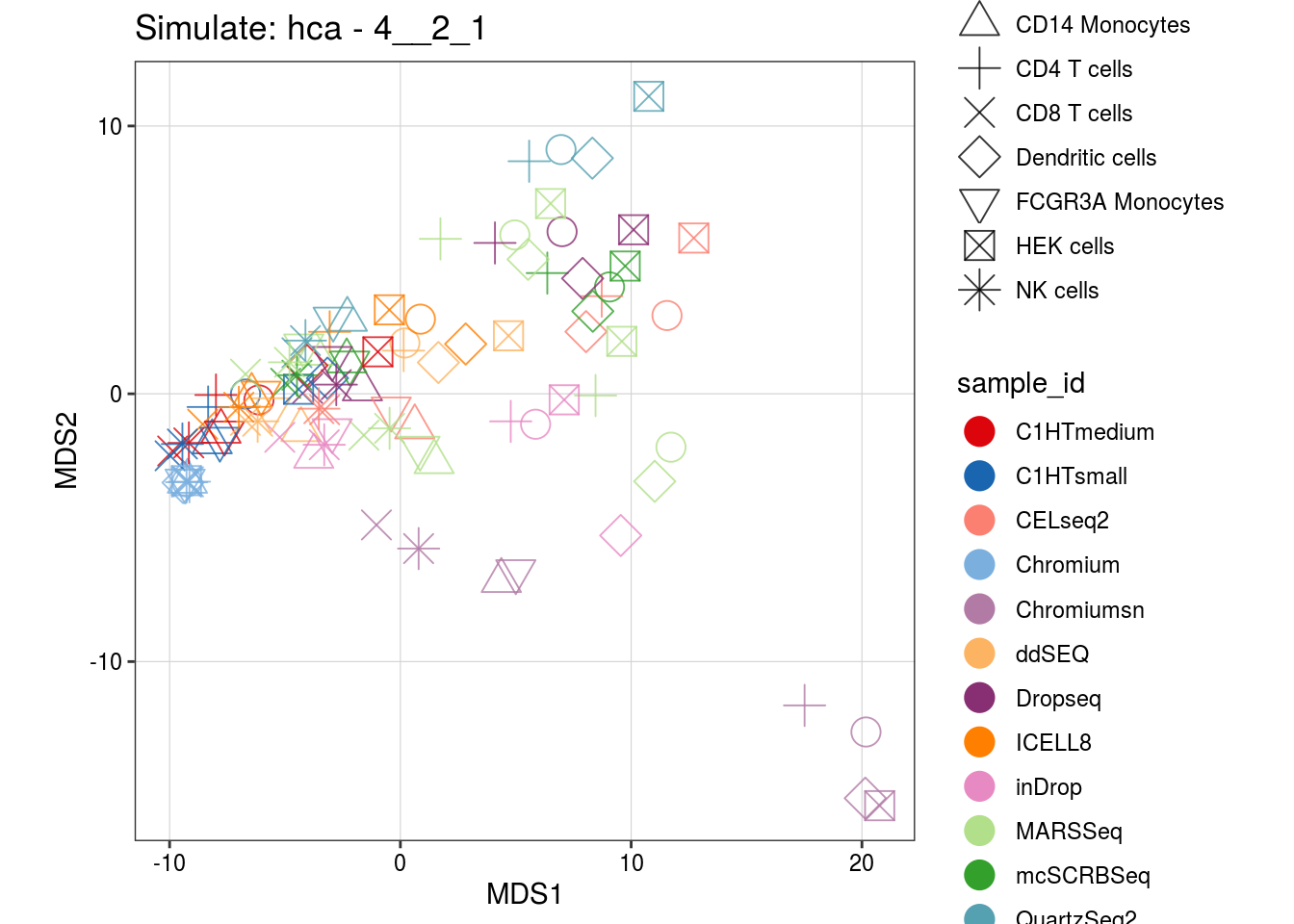
1.2__1_0

1.5__1_0

0.6__2_1
0.4__1_0
1.5__1
1__2_1

0.7__1

0.6__1
0.9__1

0.3__2_1

0.4__1

Real data
# to make aggregate functions work
sce_real$batch_id <- as.factor(colData(sce_real)[, batch_real])
sce_real$sample_id <- as.factor(colData(sce_real)[, sample_real])
sce_real$cluster_id <- as.factor(colData(sce_real)[, cluster_real])
sce_real$group_id <- as.factor(rep("A", ncol(sce_real)))
metadata(sce_real)$experiment_info <- colData(sce_real)[, c("batch_id", "cluster_id", "group_id", "sample_id")]
visTSNE(sce_real, title = "Real dataset", ids = "batch_id")## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.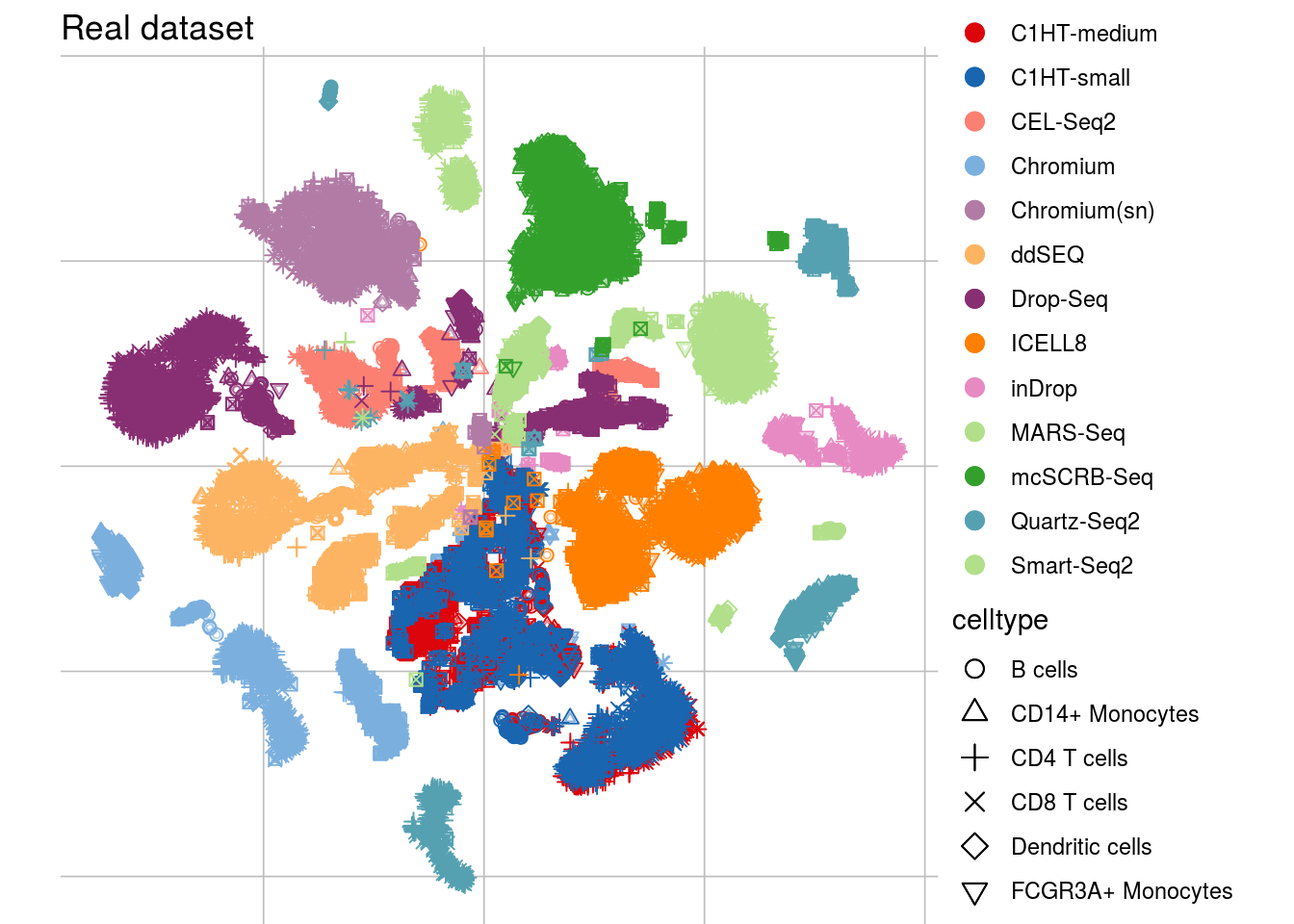
visUMAP(sce_real, title = "Real dataset", ids = "batch_id")## Scale for 'colour' is already present. Adding another scale for 'colour',
## which will replace the existing scale.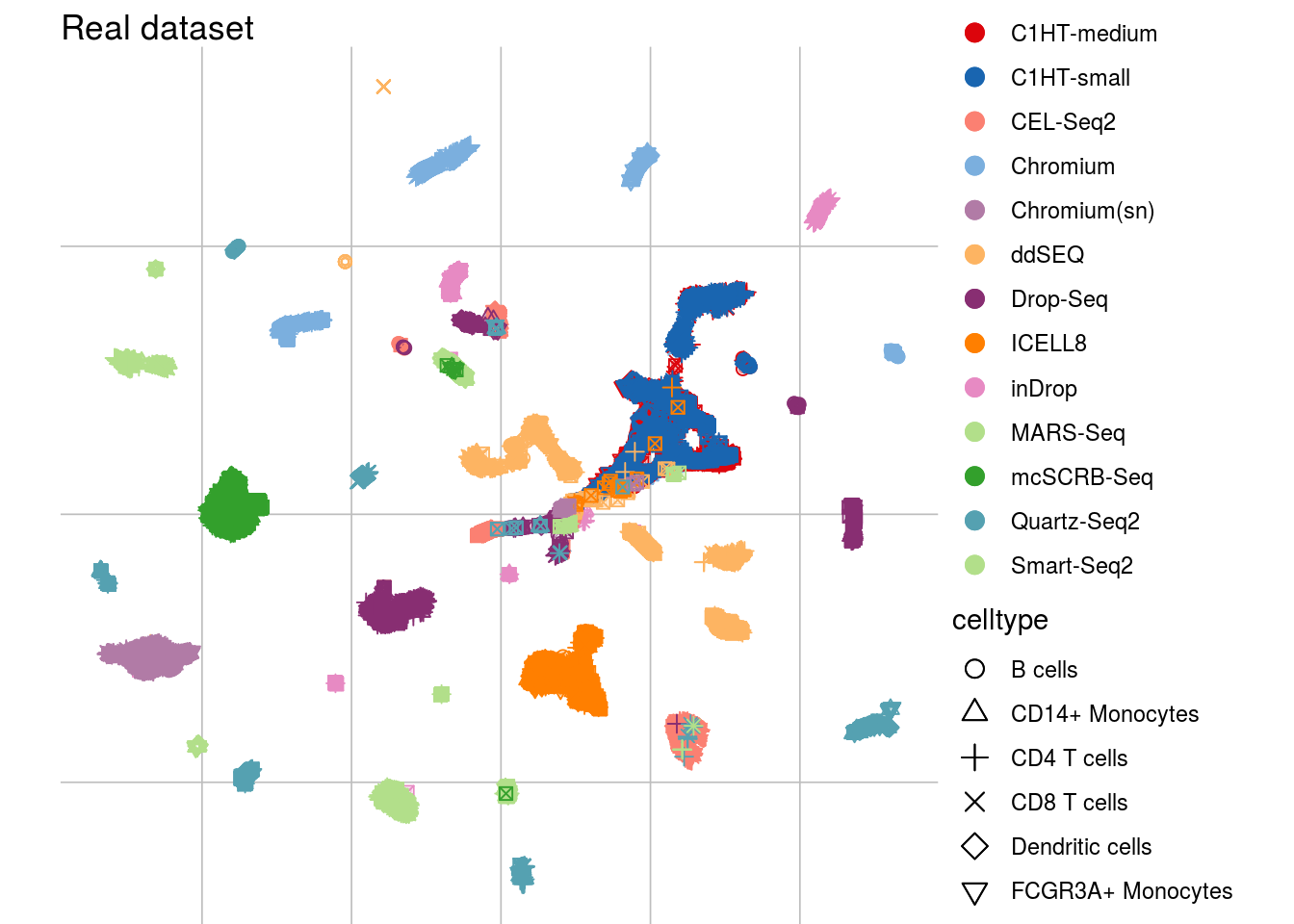
visMDS(sce_real, title = "Real dataset", ids = "batch_id")
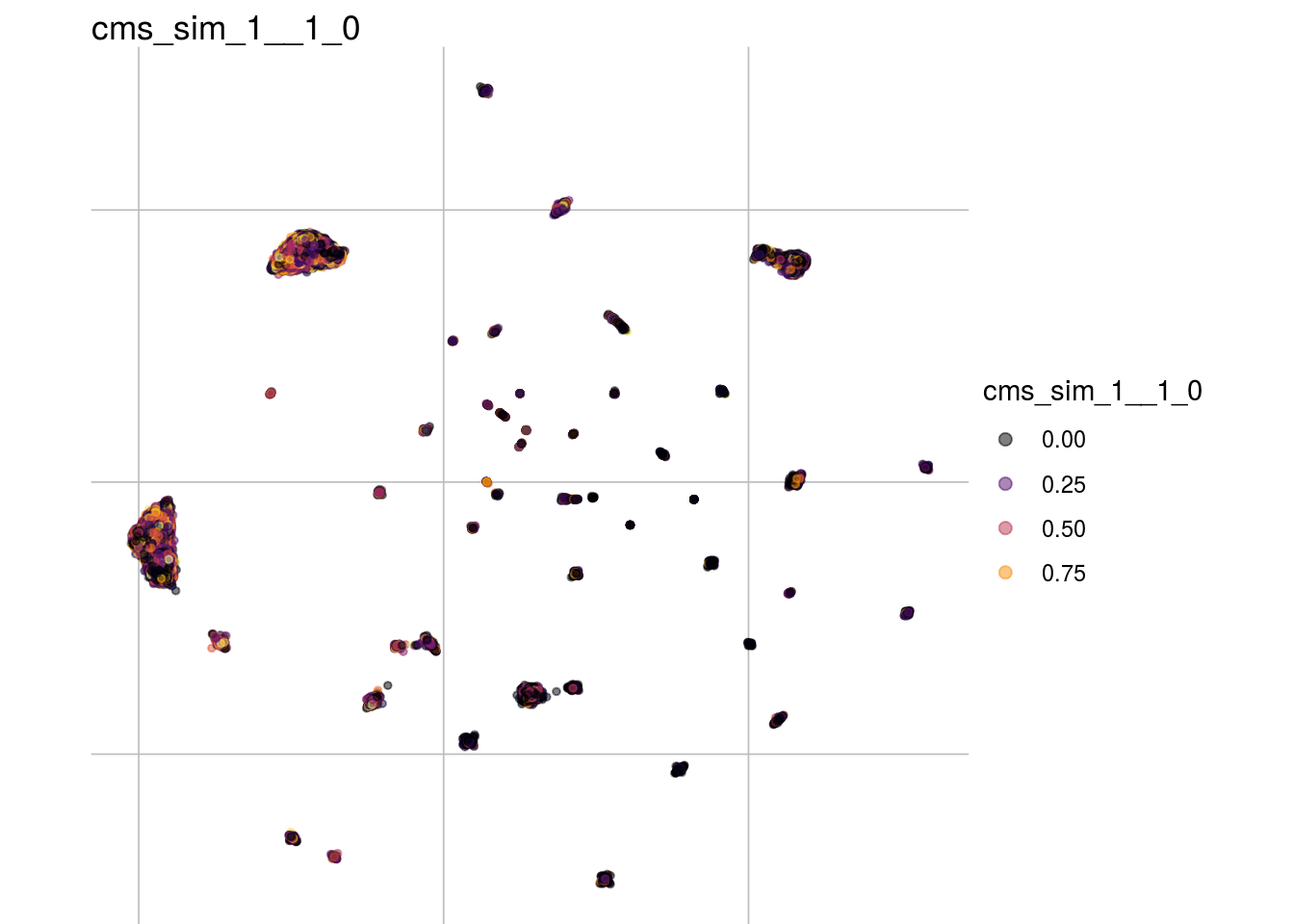
Cellspecific Mixing score simulations
cms_sim_1__1_0
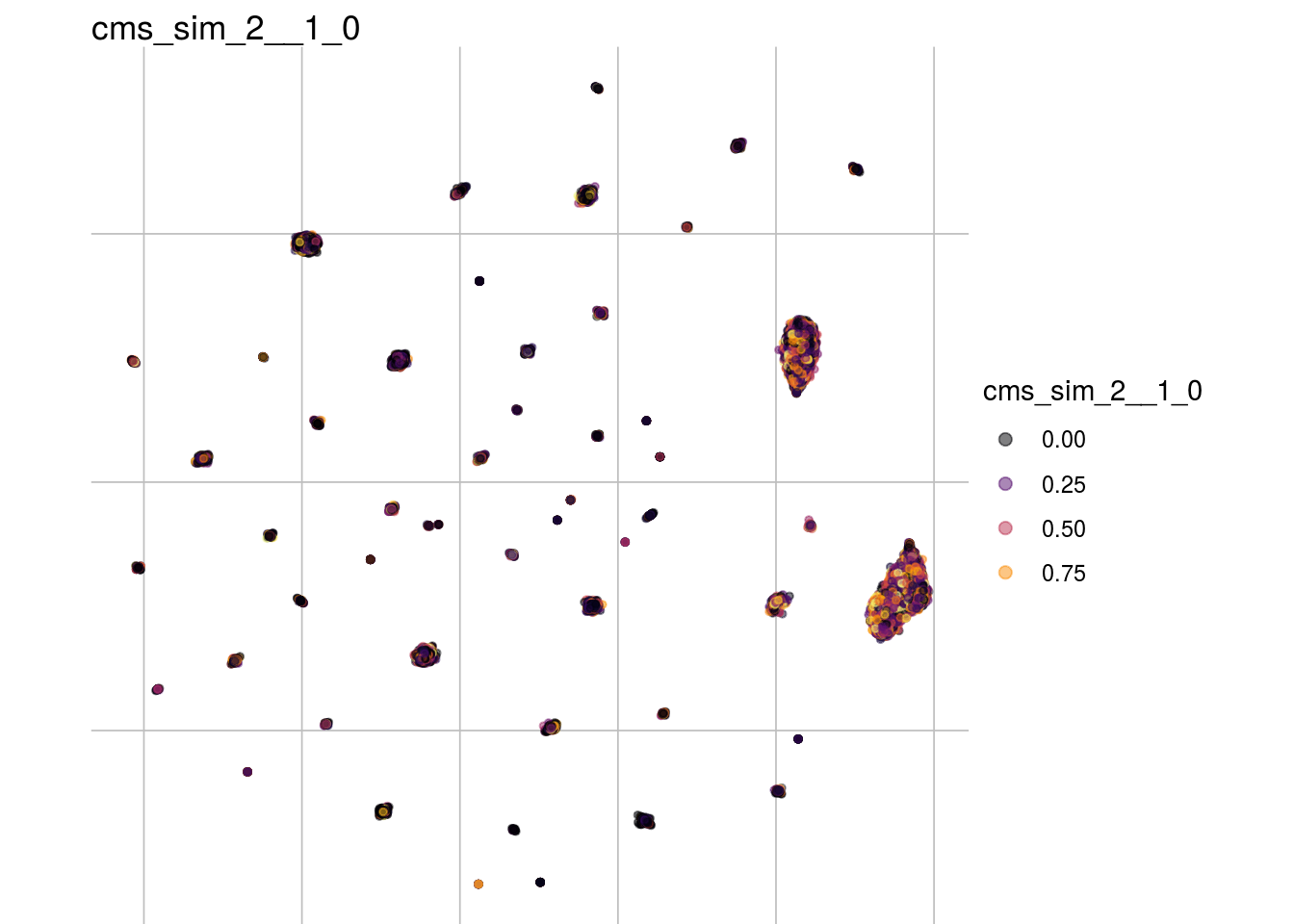
cms_sim_2__1_0
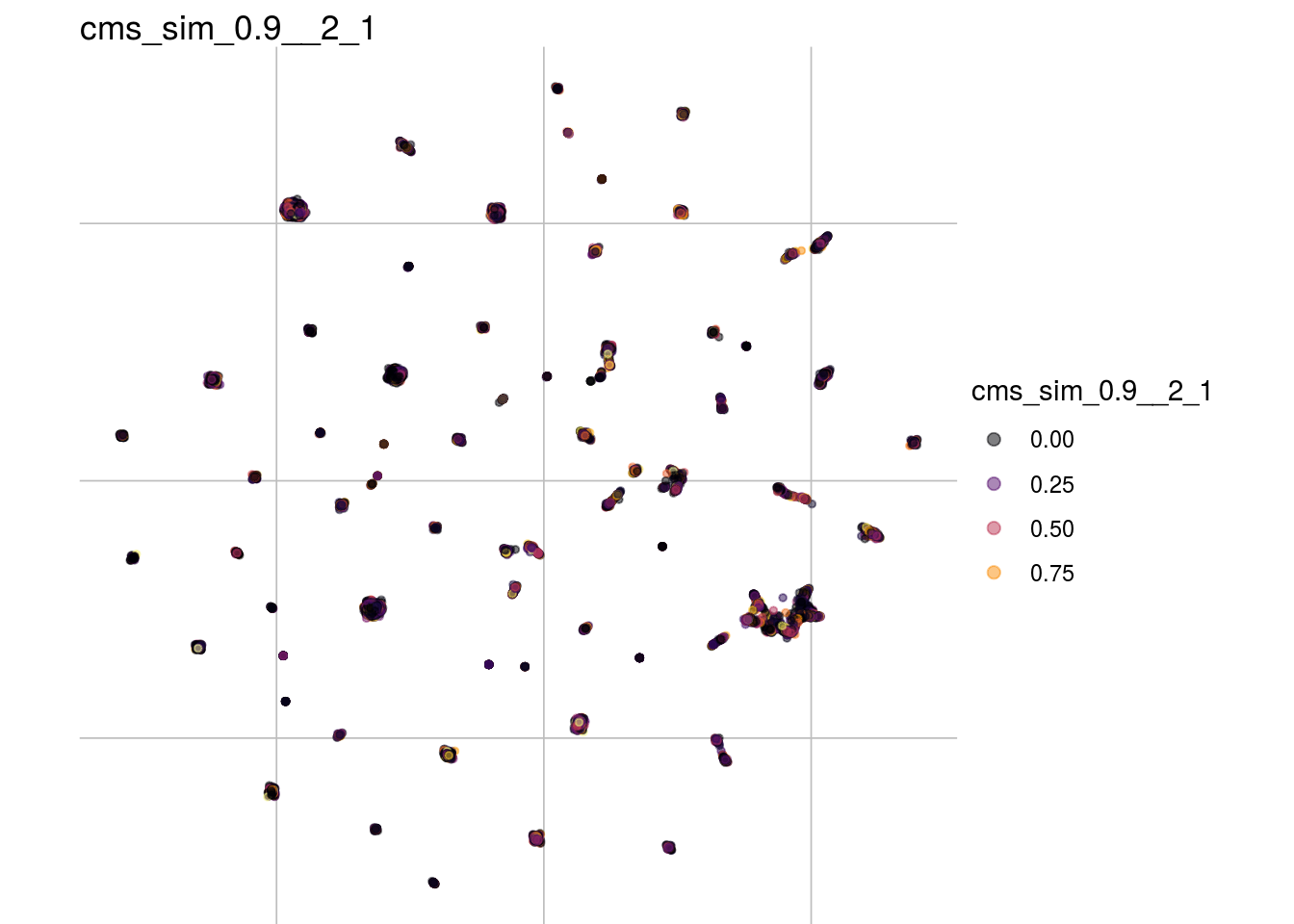
cms_sim_0.9__2_1
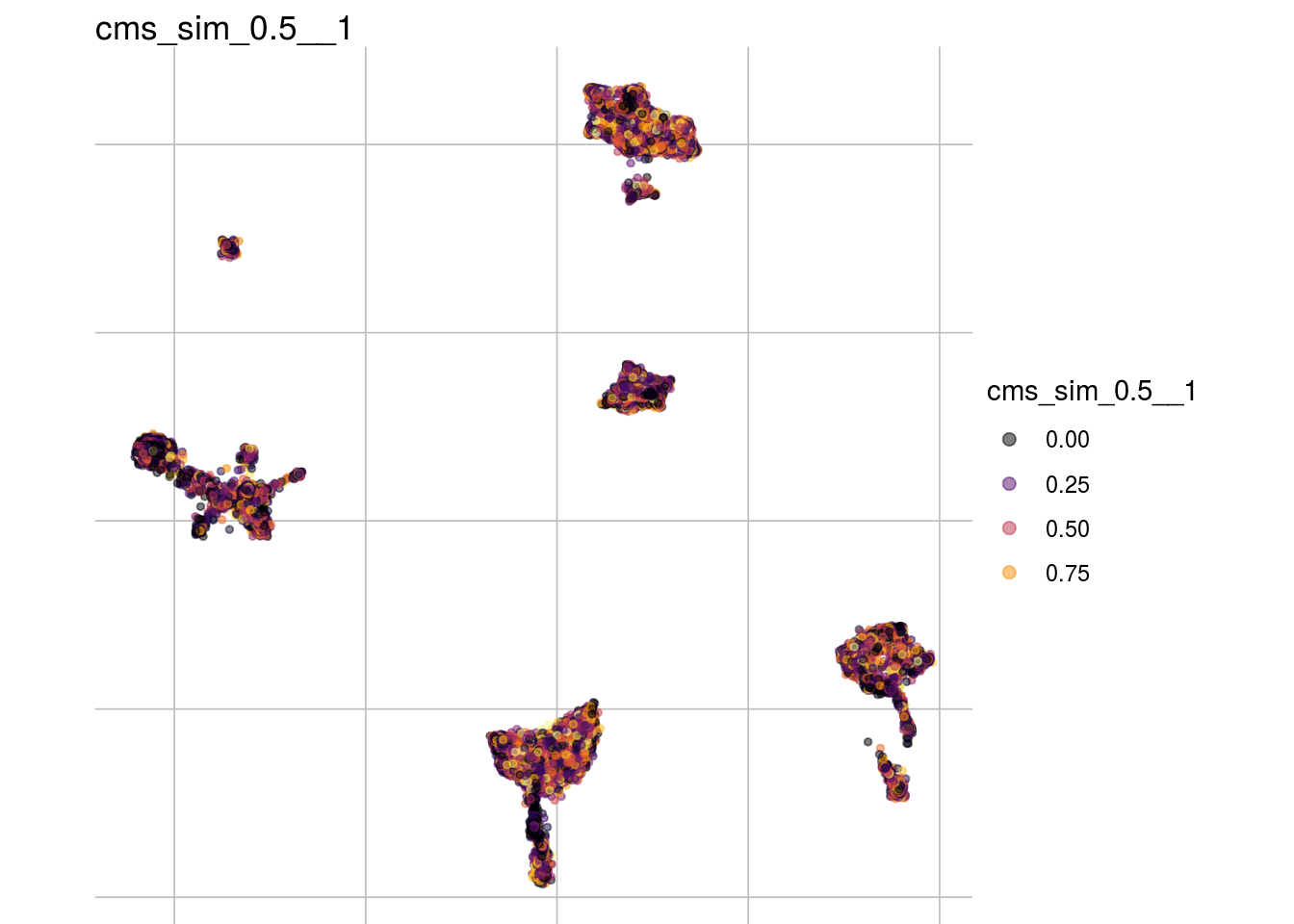
cms_sim_0.5__1
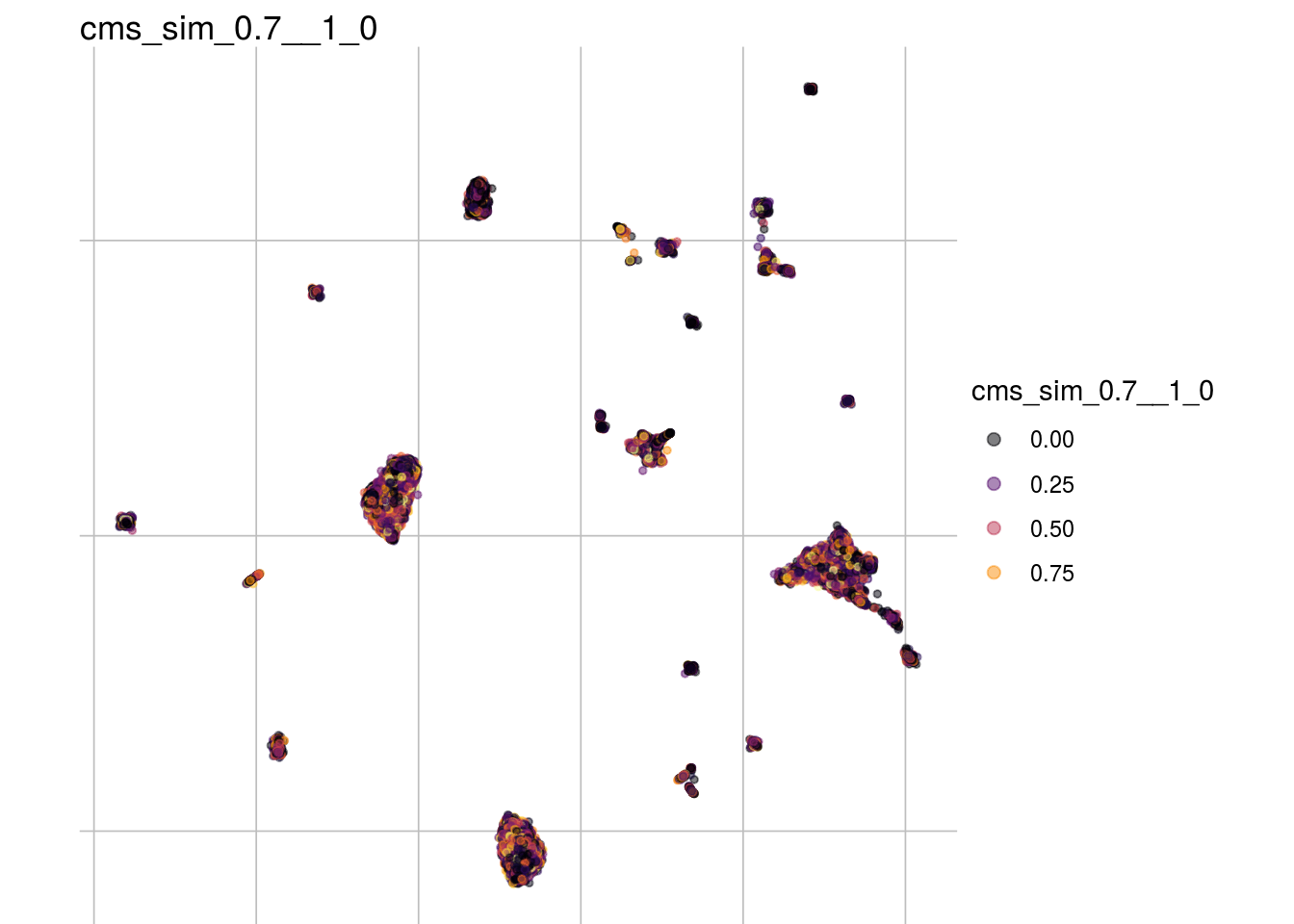
cms_sim_0.7__1_0
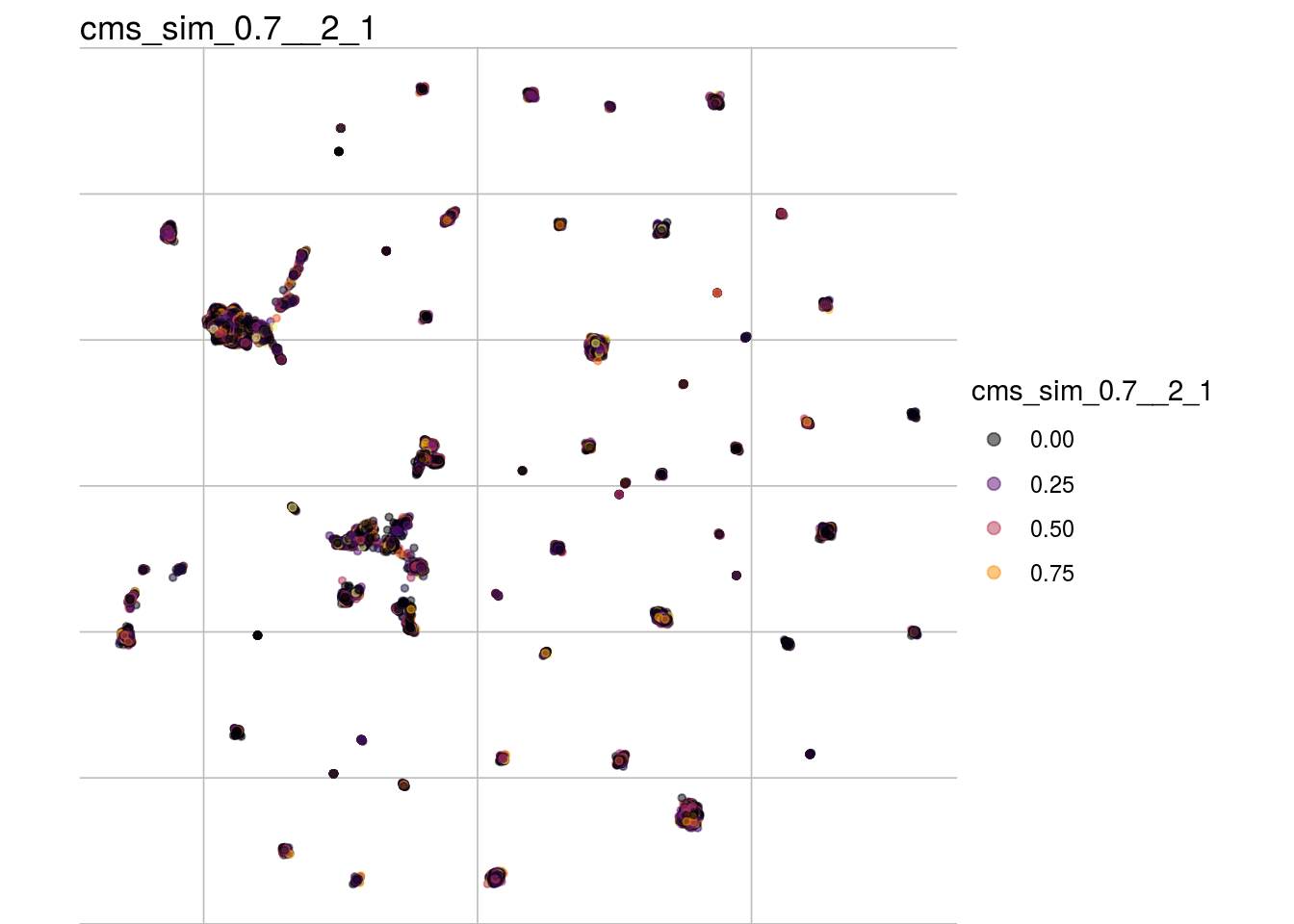
cms_sim_0.7__2_1
cms_sim_0.5__2_1
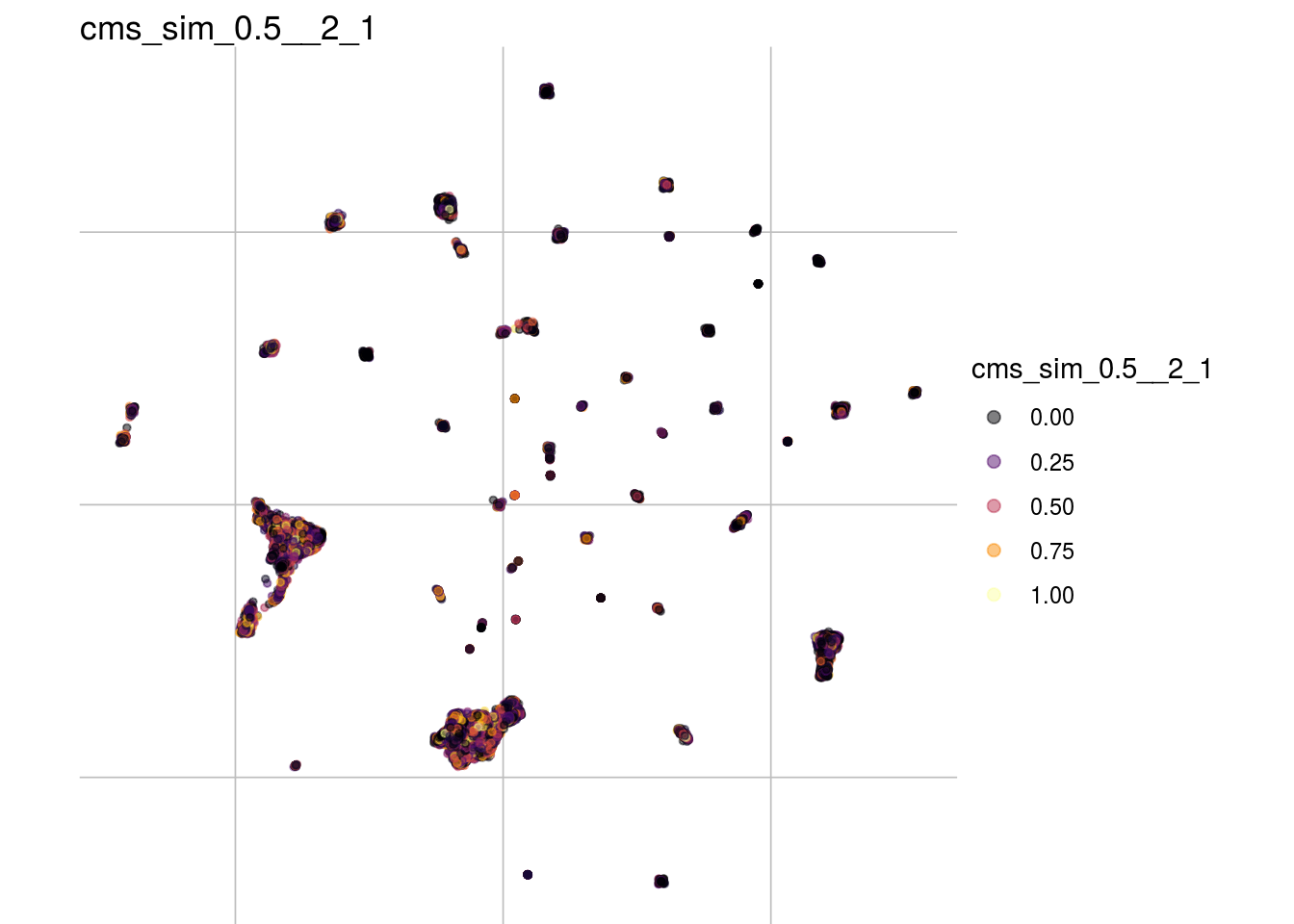
cms_sim_0.5__1_0
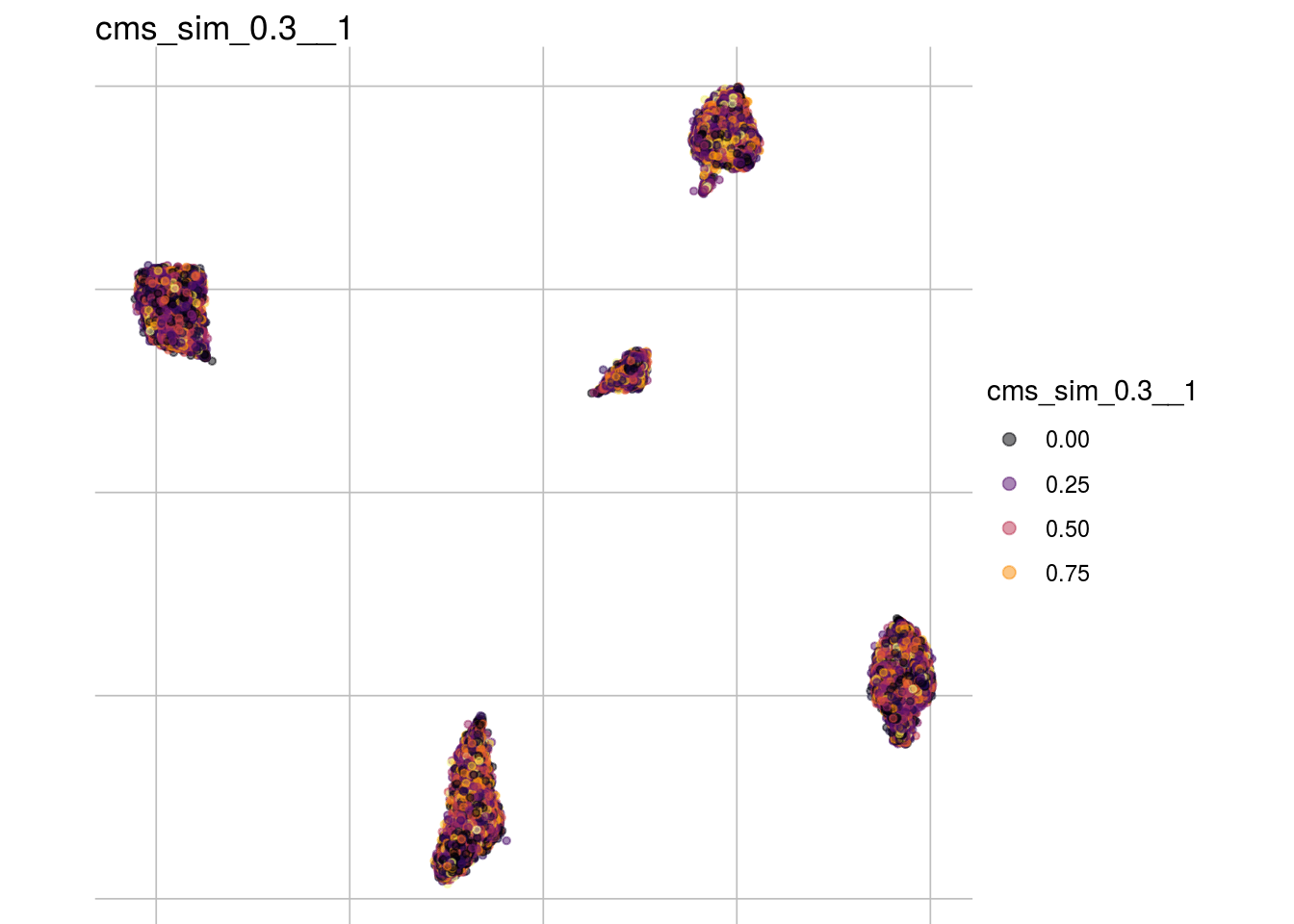
cms_sim_0.3__1
cms_sim_1.2__1

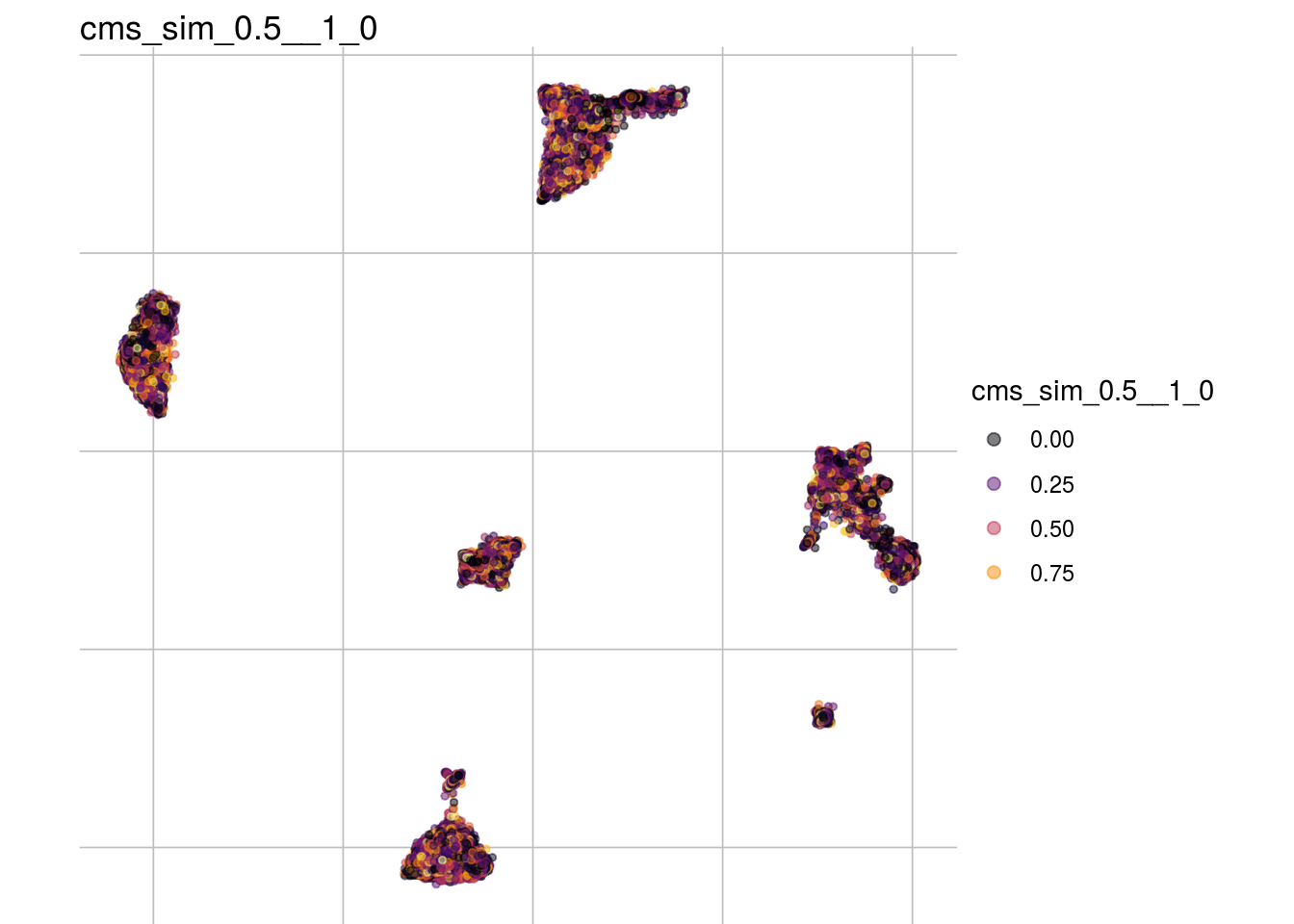
cms_sim_0.8__1_0
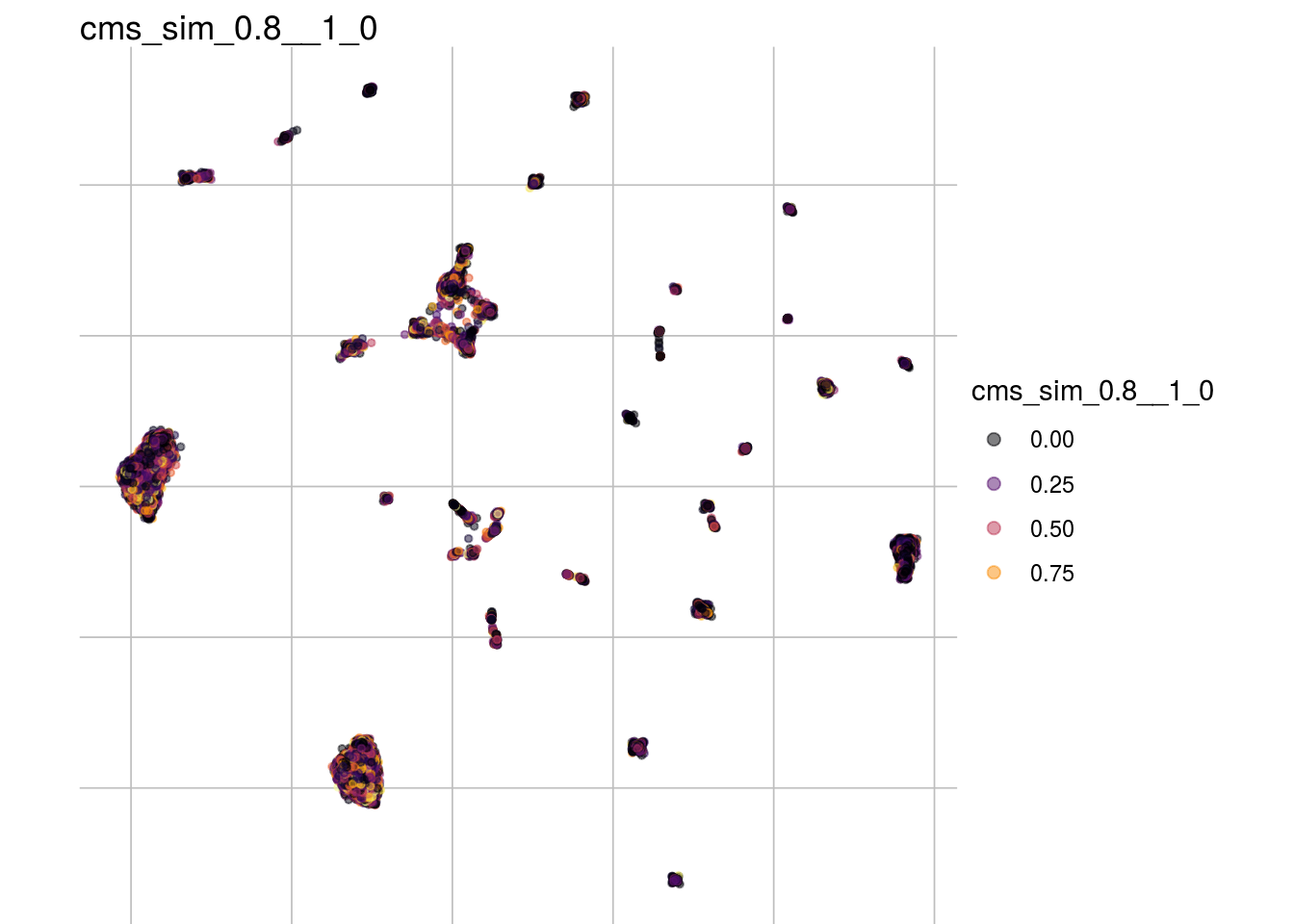
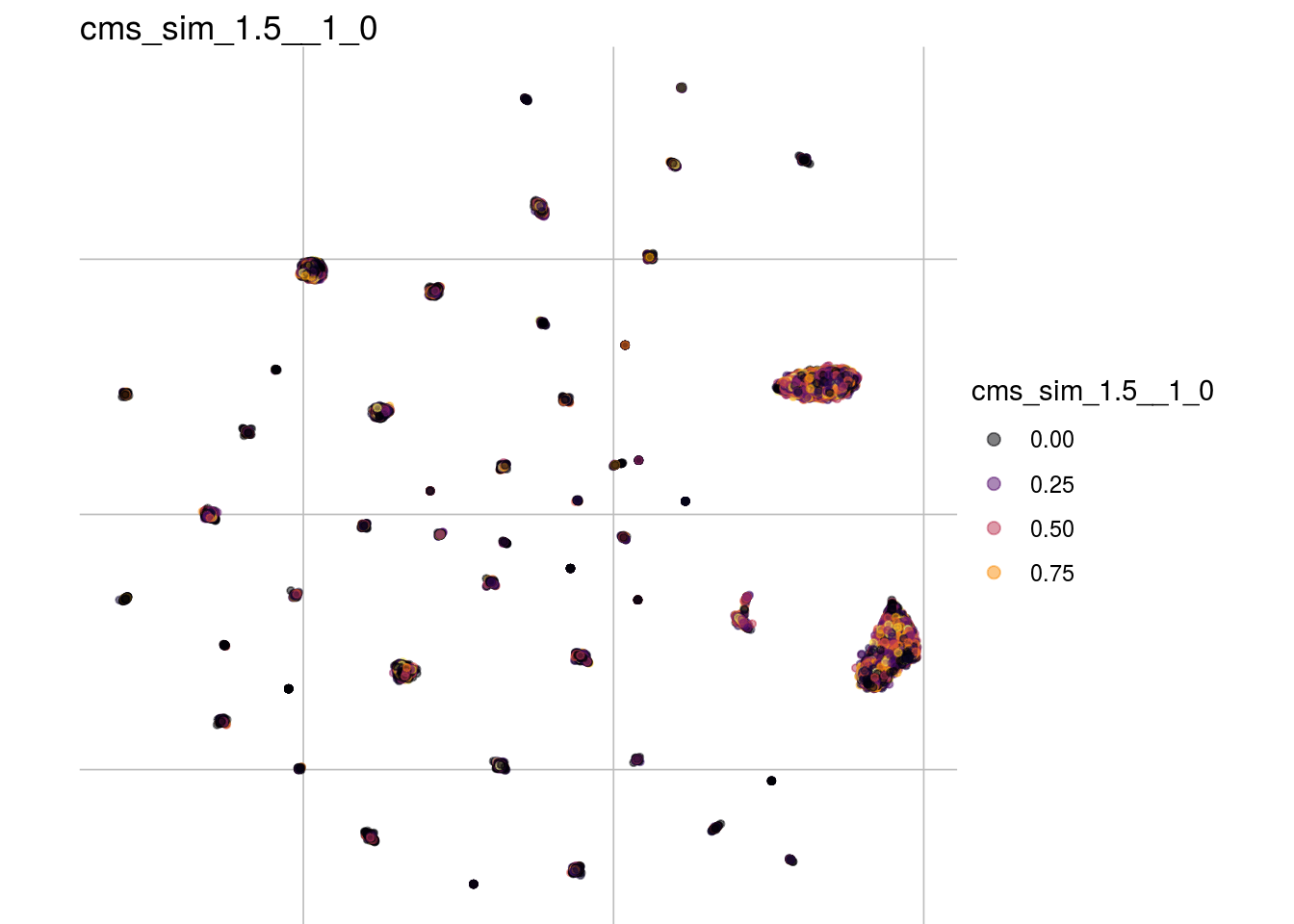
cms_sim_0.6__1_0
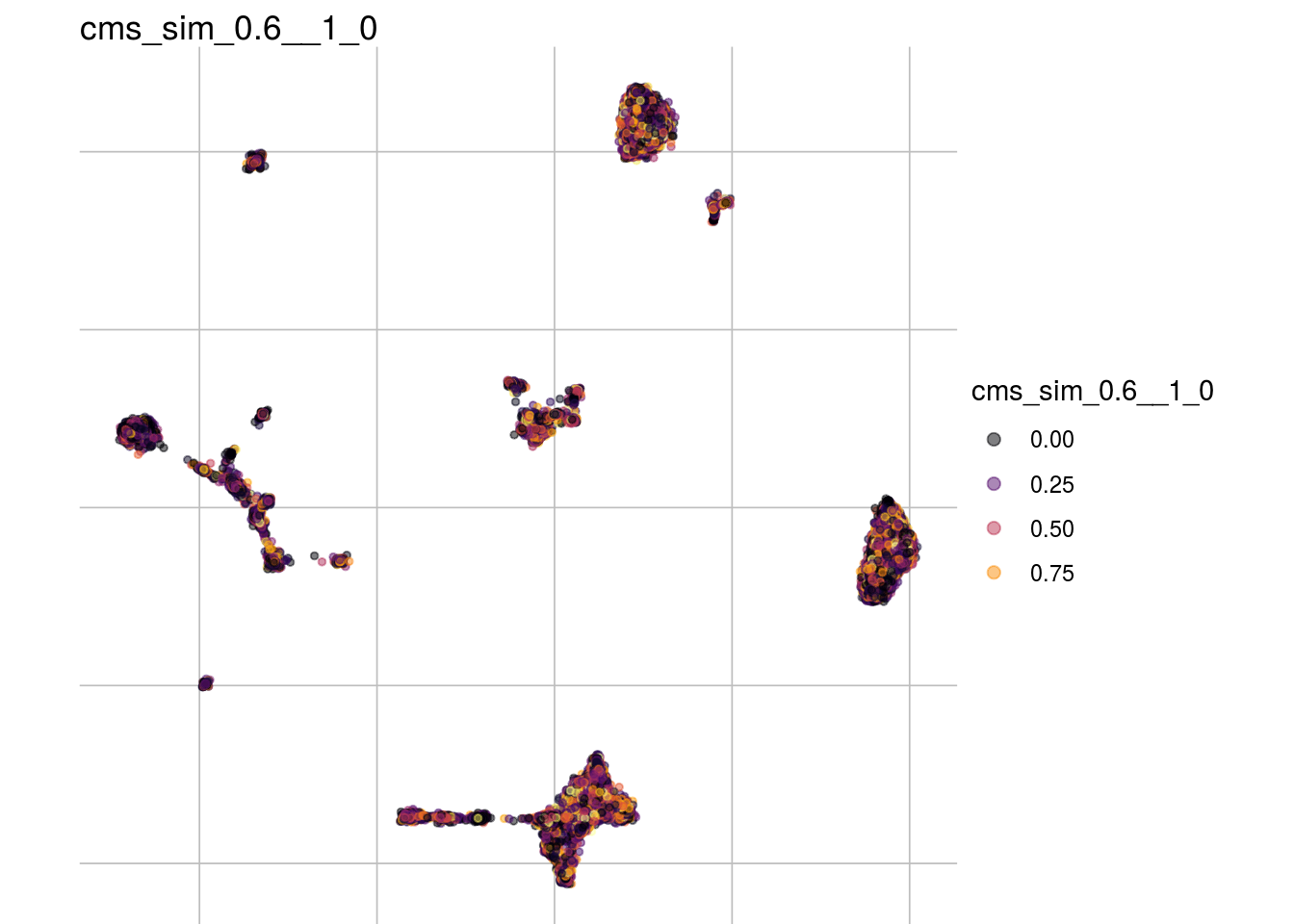
cms_sim_1__1

cms_sim_1.2__2_1

cms_sim_0.8__2_1

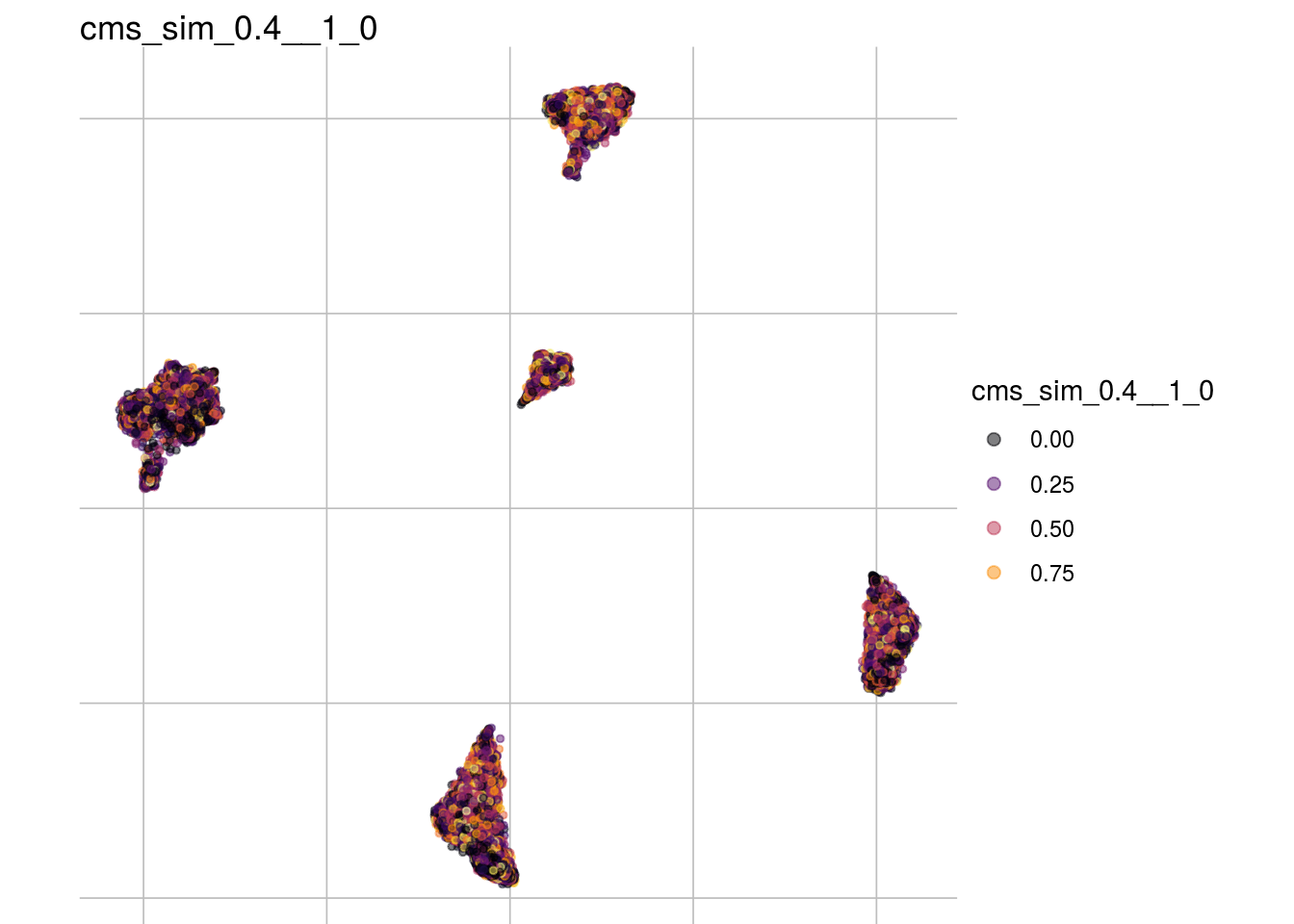
cms_sim_0.3__1_0

cms_sim_0.4__2_1

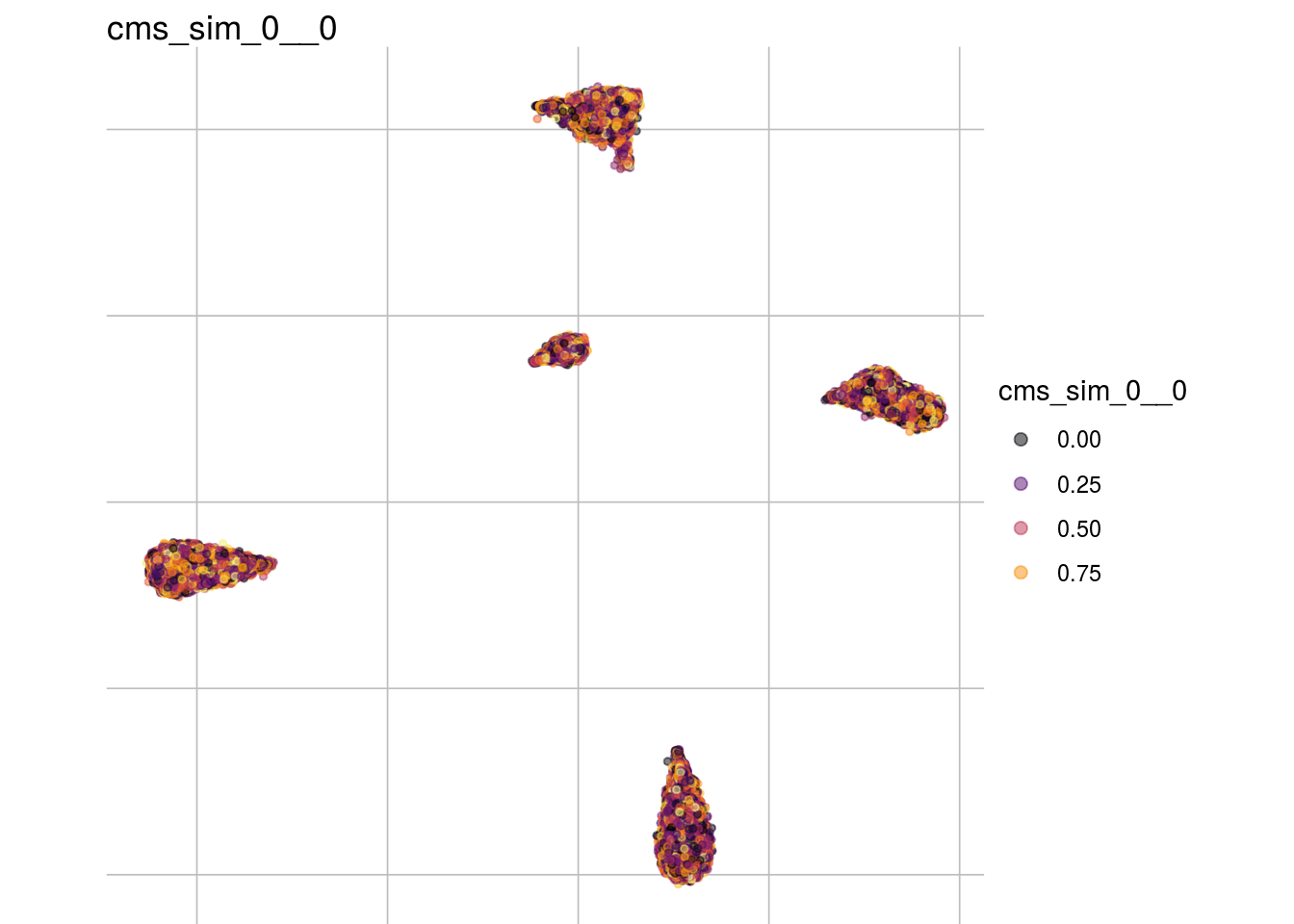
cms_sim_0__0
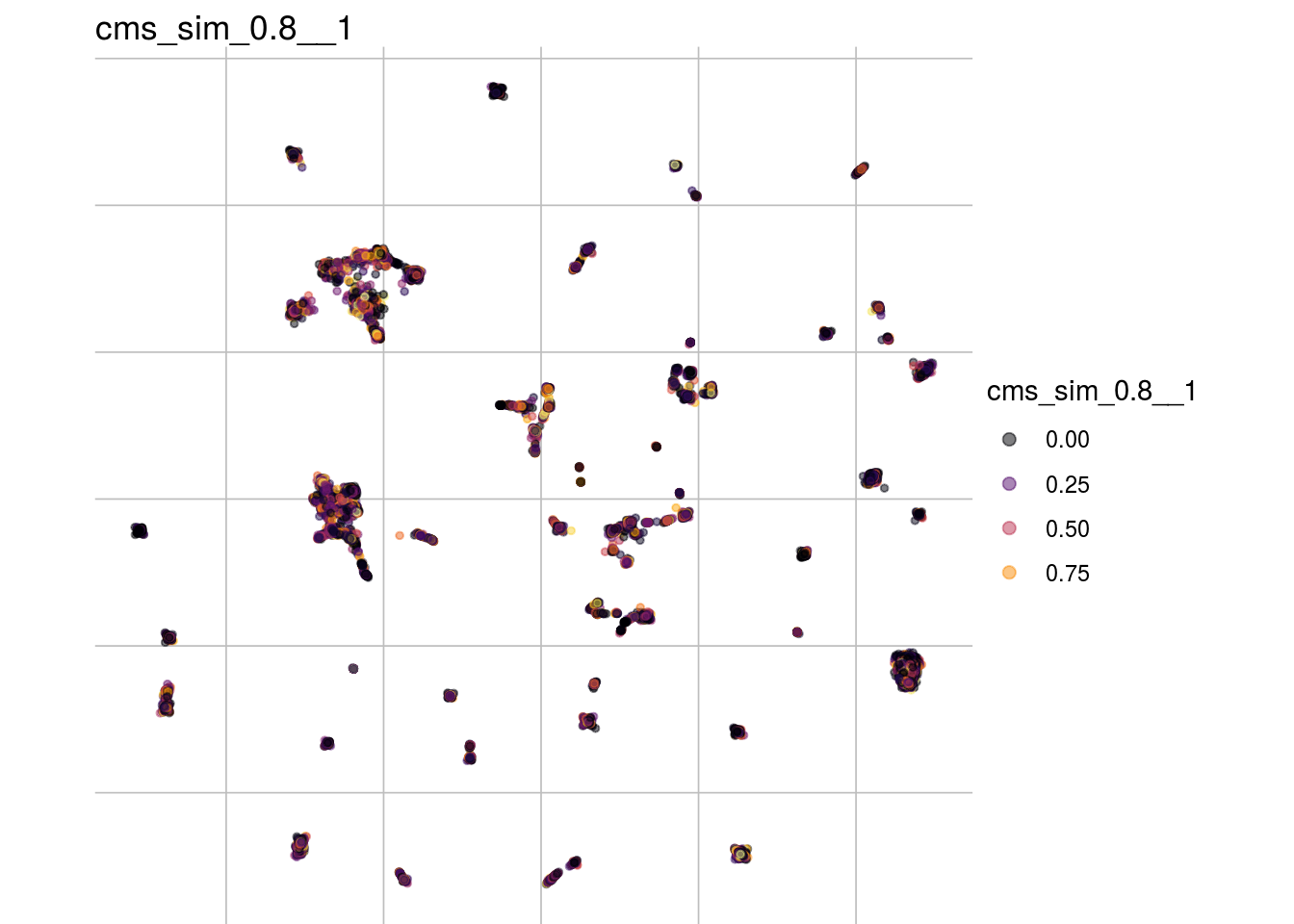
cms_sim_0.8__1
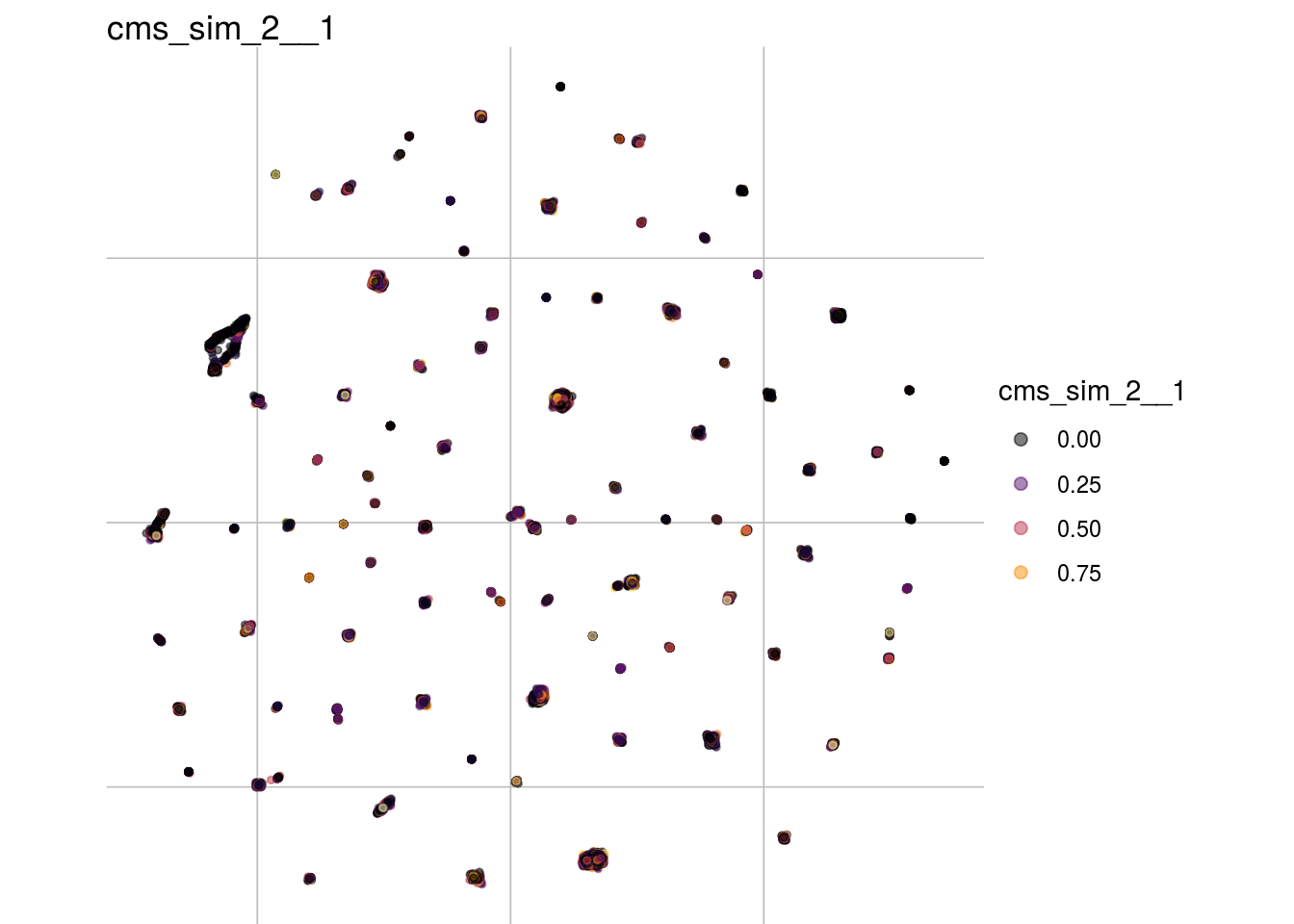
cms_sim_2__1
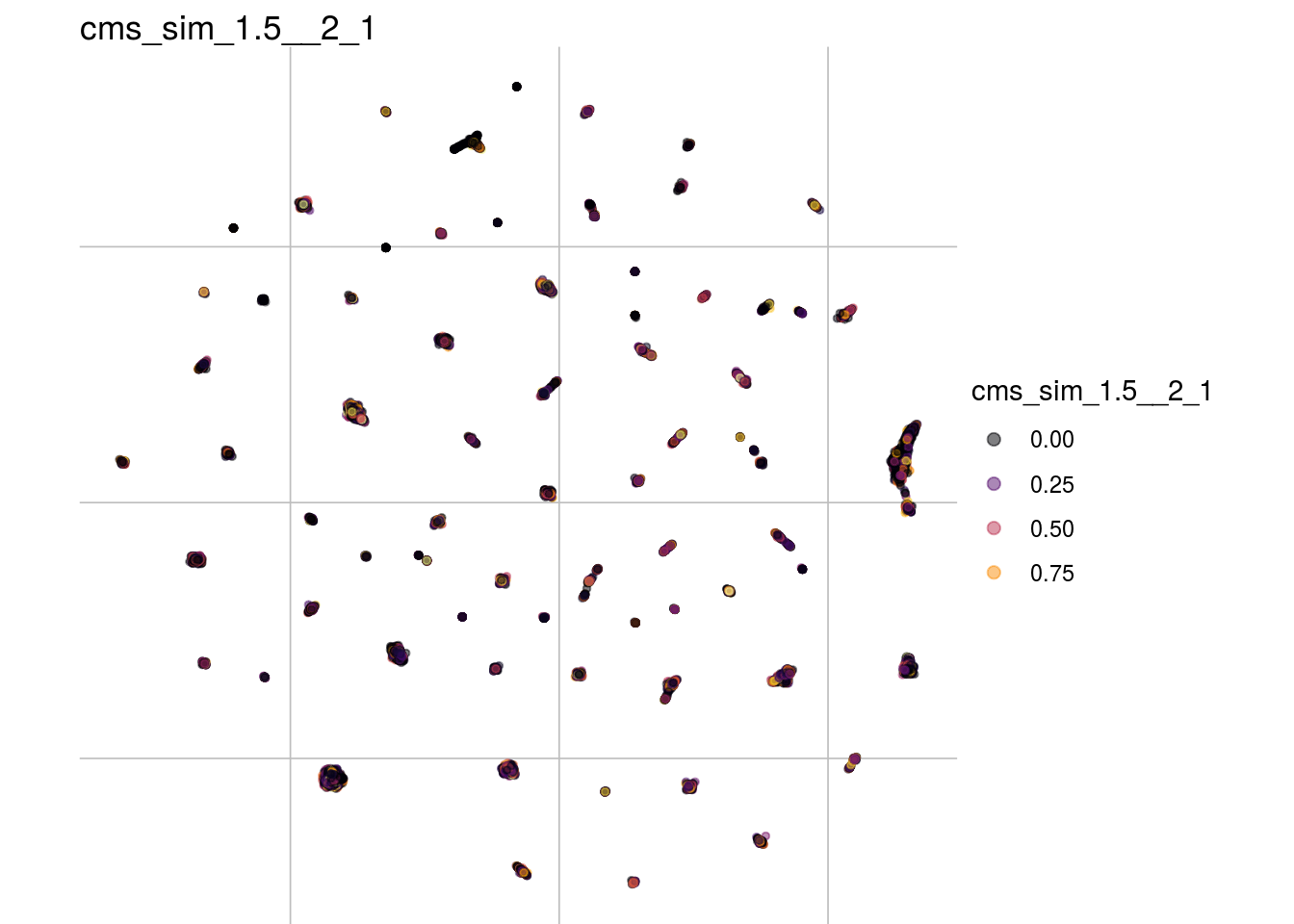
cms_sim_1.5__2_1
cms_sim_0.9__1_0

cms_sim_1.2__1_0

cms_sim_1.5__1_0
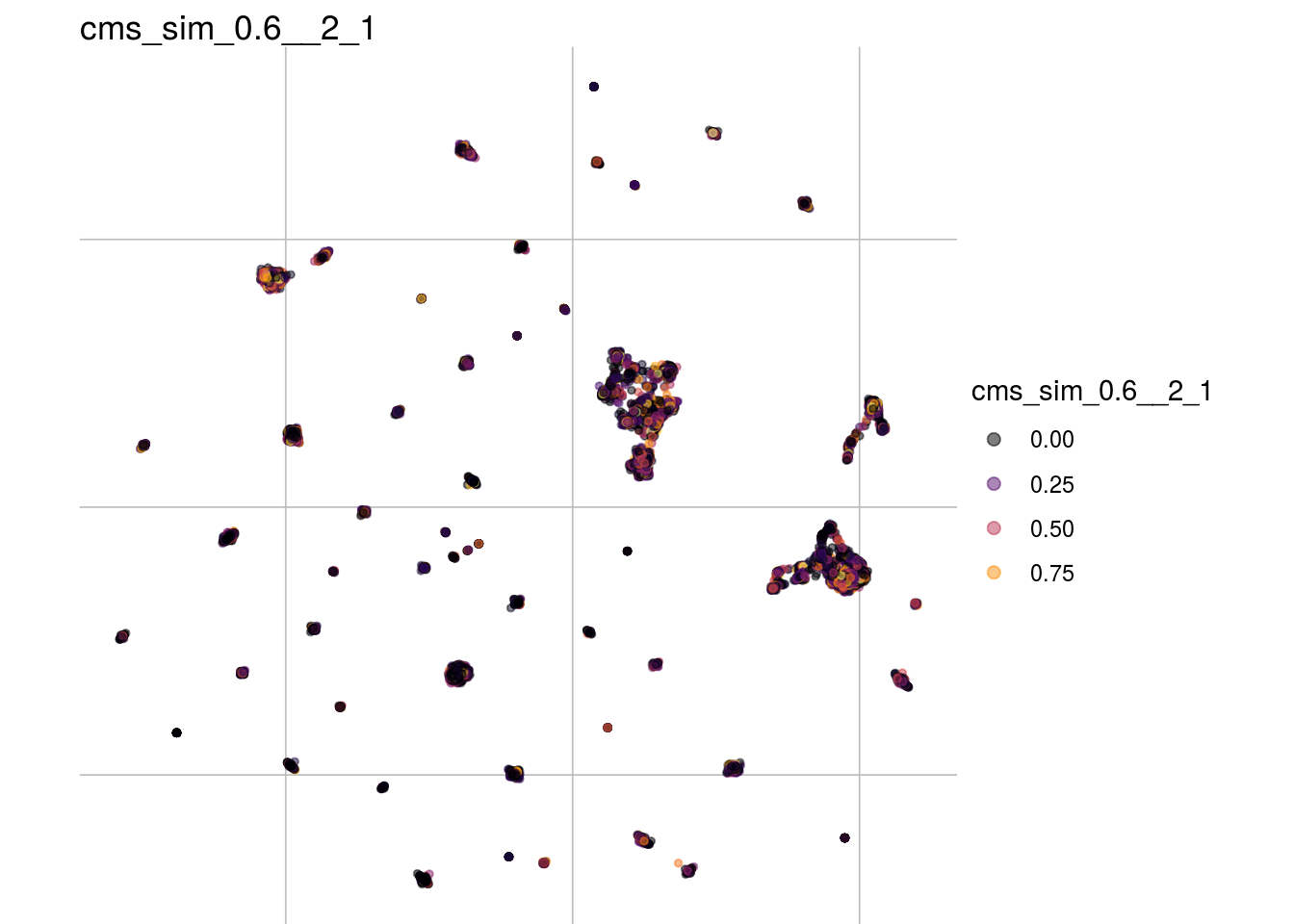
cms_sim_0.6__2_1
cms_sim_0.4__1_0
cms_sim_1.5__1
cms_sim_1__2_1
cms_sim_0.7__1

cms_sim_0.6__1
cms_sim_0.9__1
cms_sim_0.3__2_1
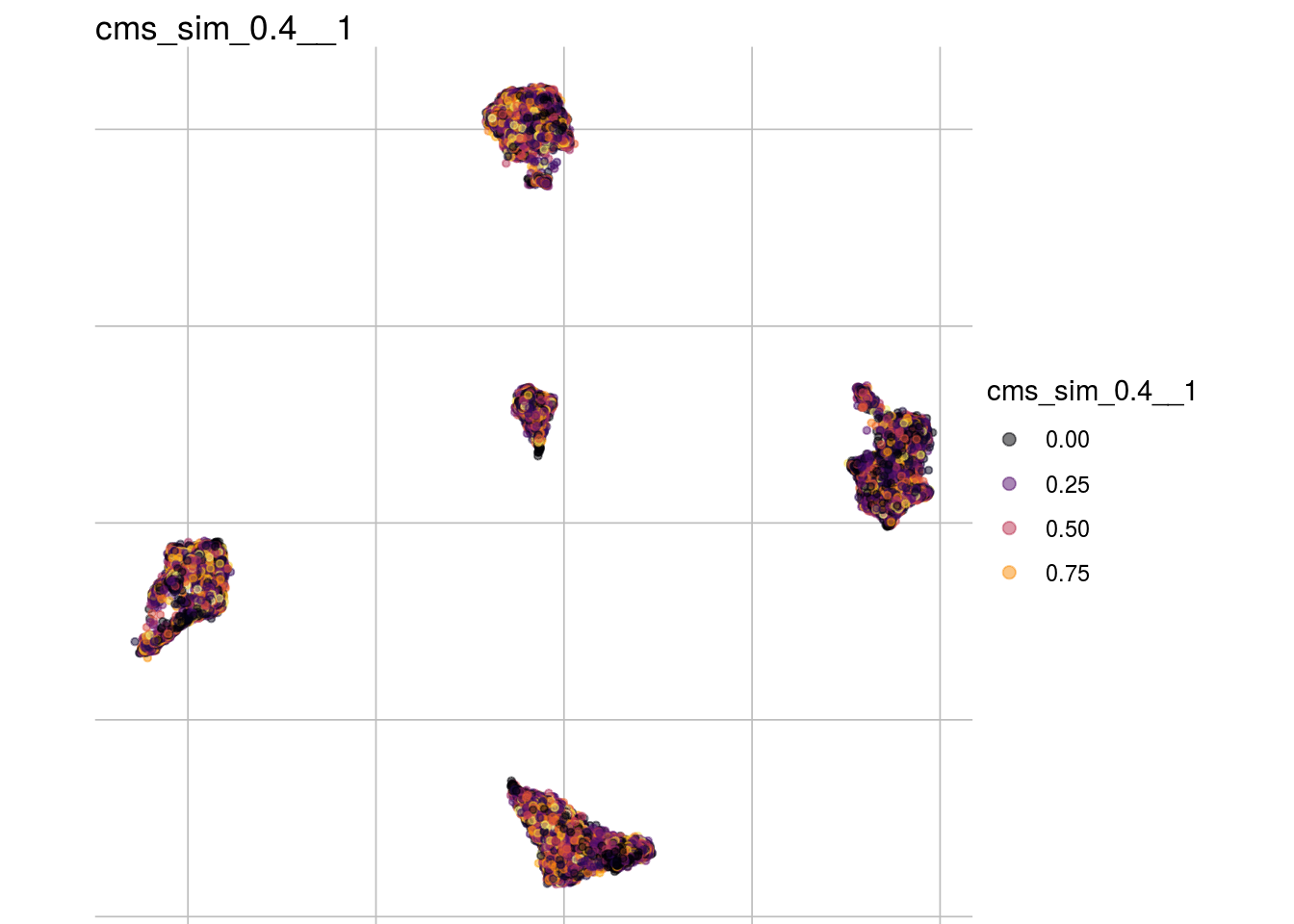
cms_sim_0.4__1