# install.packages("dplyr")
# install.packages("ggplot2")
# install.packages("SingleCellExperiment") #failed
# install.packages("scater") #failed
# install.packages("scran") #failed
# install.packages("UpSetR")
# install.packages("googlesheets4")
# install.packages("cowplot")
# install.packages("BiocParallel") #failed
# install.packages("scDblFinder") #failed
# install.packages("tibble")
# install.packages("ggpubr") #failed
# install.packages("reshape2")
# install.packages("pheatmap")
# install.packages("RColorBrewer")
# install.packages("batchelor") #failed# install.packages('BiocManager')# BiocManager::install("SingleCellExperiment")
#BiocManager::install("scater")
# BiocManager::install("scran")
# BiocManager::install("BiocParallel")
# BiocManager::install("scDblFinder")
# BiocManager::install("ggpubr")
# BiocManager::install("batchelor")
# BiocManager::install("DropletUtils")# install.packages("knitr")require("knitr")
knitr::opts_chunk$set(fig.width=10, fig.height=12)Caricamento del pacchetto richiesto: knitr
suppressPackageStartupMessages({
library(dplyr)
library(ggplot2)
library(SingleCellExperiment)
library(scater)
library(scran)
library(UpSetR)
library(googlesheets4)
library(cowplot)
library(BiocParallel)
library(scDblFinder)
library(tibble)
library(ggpubr)
library(reshape2)
library(pheatmap)
library(RColorBrewer)
library(batchelor)
library(DropletUtils)
})Warning message:
“il pacchetto ‘UpSetR’ è stato creato con R versione 4.3.0”
Warning message:
“il pacchetto ‘googlesheets4’ è stato creato con R versione 4.3.0”
Warning message:
“il pacchetto ‘batchelor’ è stato creato con R versione 4.3.0”
Warning message:
“il pacchetto ‘DropletUtils’ è stato creato con R versione 4.3.0”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::.__C__DummyArrayViewport’ con ‘DelayedArray::.__C__DummyArrayViewport’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::DummyArrayGrid’ con ‘DelayedArray::DummyArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::DummyArrayViewport’ con ‘DelayedArray::DummyArrayViewport’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::write_block’ con ‘DelayedArray::write_block’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::maxlength’ con ‘DelayedArray::maxlength’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::arbind’ con ‘DelayedArray::arbind’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::aperm.ArbitraryArrayGrid’ con ‘DelayedArray::aperm.ArbitraryArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::refdim’ con ‘DelayedArray::refdim’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::downsample’ con ‘DelayedArray::downsample’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.character.Array’ con ‘DelayedArray::as.character.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::mapToGrid’ con ‘DelayedArray::mapToGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.complex.Array’ con ‘DelayedArray::as.complex.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::Mindex2Lindex’ con ‘DelayedArray::Mindex2Lindex’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.matrix.Array’ con ‘DelayedArray::as.matrix.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::is_sparse’ con ‘DelayedArray::is_sparse’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::.__C__RegularArrayGrid’ con ‘DelayedArray::.__C__RegularArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::.__C__SafeArrayViewport’ con ‘DelayedArray::.__C__SafeArrayViewport’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::.__C__Array’ con ‘DelayedArray::.__C__Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::.__C__ArrayViewport’ con ‘DelayedArray::.__C__ArrayViewport’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::RegularArrayGrid’ con ‘DelayedArray::RegularArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::makeNindexFromArrayViewport’ con ‘DelayedArray::makeNindexFromArrayViewport’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::extract_array’ con ‘DelayedArray::extract_array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::.__C__ArbitraryArrayGrid’ con ‘DelayedArray::.__C__ArbitraryArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::aperm.DummyArrayGrid’ con ‘DelayedArray::aperm.DummyArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.array.Array’ con ‘DelayedArray::as.array.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.integer.Array’ con ‘DelayedArray::as.integer.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::is_sparse<-’ con ‘DelayedArray::is_sparse<-’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.raw.Array’ con ‘DelayedArray::as.raw.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::.__C__DummyArrayGrid’ con ‘DelayedArray::.__C__DummyArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::ArbitraryArrayGrid’ con ‘DelayedArray::ArbitraryArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.character.ArrayGrid’ con ‘DelayedArray::as.character.ArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.numeric.Array’ con ‘DelayedArray::as.numeric.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::ArrayViewport’ con ‘DelayedArray::ArrayViewport’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::acbind’ con ‘DelayedArray::acbind’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::t.Array’ con ‘DelayedArray::t.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::read_block’ con ‘DelayedArray::read_block’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::rowsum’ con ‘DelayedArray::rowsum’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::.__C__ArrayGrid’ con ‘DelayedArray::.__C__ArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::aperm.RegularArrayGrid’ con ‘DelayedArray::aperm.RegularArrayGrid’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.vector.Array’ con ‘DelayedArray::as.vector.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::Lindex2Mindex’ con ‘DelayedArray::Lindex2Mindex’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::colsum’ con ‘DelayedArray::colsum’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::mapToRef’ con ‘DelayedArray::mapToRef’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.data.frame.Array’ con ‘DelayedArray::as.data.frame.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“sostituzione dell'importazione precedente ‘S4Arrays::as.logical.Array’ con ‘DelayedArray::as.logical.Array’ durante il caricamento di ‘HDF5Array’”
Warning message:
“trovate tabelle metodi multiple per ‘extract_array’”
Warning message:
“trovate tabelle metodi multiple per ‘is_sparse’”
Warning message:
“trovate tabelle metodi multiple per ‘is_sparse<-’”
Warning message:
“trovate tabelle metodi multiple per ‘write_block’”datadir <- dir("../data", "filtered_feature_bc_matrix$",
full.names = TRUE, recursive = TRUE,
include.dirs = TRUE)
names(datadir) <- sapply(strsplit(datadir, "/"), .subset, 3)
datadir- donor1_fat
- '../data/donor1_fat/count/sample_filtered_feature_bc_matrix'
- donor1_skin
- '../data/donor1_skin/count/sample_filtered_feature_bc_matrix'
- donor2_fat
- '../data/donor2_fat/count/sample_filtered_feature_bc_matrix'
- donor3_fat
- '../data/donor3_fat/count/sample_filtered_feature_bc_matrix'
- donor3_skin
- '../data/donor3_skin/count/sample_filtered_feature_bc_matrix'
- donor4_fat
- '../data/donor4_fat/count/sample_filtered_feature_bc_matrix'
- donor4_skin
- '../data/donor4_skin/count/sample_filtered_feature_bc_matrix'
- donor5_fat
- '../data/donor5_fat/count/sample_filtered_feature_bc_matrix'
- donor5_recovery
- '../data/donor5_recovery/filtered_feature_bc_matrix'
- donor5_skin
- '../data/donor5_skin/count/sample_filtered_feature_bc_matrix'
- donor6_fat
- '../data/donor6_fat/count/sample_filtered_feature_bc_matrix'
- donor6_skin
- '../data/donor6_skin/count/sample_filtered_feature_bc_matrix'
- donor7_fat
- '../data/donor7_fat/count/sample_filtered_feature_bc_matrix'
- donor7_skin
- '../data/donor7_skin/count/sample_filtered_feature_bc_matrix'
stub <- "https://docs.google.com/spreadsheets/d/"
md_sheet <- paste0(stub, "1yEqqOHX7TY1A-lmEQu3cPcFY60IconZWR6W349xH2pY")
md_sheet
'https://docs.google.com/spreadsheets/d/1yEqqOHX7TY1A-lmEQu3cPcFY60IconZWR6W349xH2pY'
googlesheets4::gs4_deauth()
md <- googlesheets4::read_sheet(md_sheet, sheet = "single-cell") %>%
filter(keep == 1)
md✔ Reading from halin-bauer-hLECs-metadata.
✔ Range ''single-cell''.
| sample_name | donor | tissue | tissue_origin | sequencing_run | keep | sex | notes |
|---|---|---|---|---|---|---|---|
| <chr> | <dbl> | <chr> | <chr> | <dbl> | <dbl> | <chr> | <chr> |
| donor1_fat | 1 | fat | abdomen | 1 | 1 | F | NA |
| donor1_skin | 1 | skin | abdomen | 1 | 1 | F | NA |
| donor2_fat | 2 | fat | arm | 1 | 1 | F | NA |
| donor3_fat | 3 | fat | abdomen | 2 | 1 | F | has contamination |
| donor3_skin | 3 | skin | abdomen | 2 | 1 | F | NA |
| donor4_fat | 4 | fat | abdomen | 2 | 1 | F | NA |
| donor4_skin | 4 | skin | abdomen | 2 | 1 | F | NA |
| donor5_recovery | 5 | mixed | abdomen | 4 | 1 | ? | recovered sample |
| donor6_fat | 6 | fat | abdomen | 3 | 1 | M | NA |
| donor6_skin | 6 | skin | abdomen | 3 | 1 | M | NA |
| donor7_fat | 7 | fat | thigh | 3 | 1 | ? | has contamination |
| donor7_skin | 7 | skin | thigh | 3 | 1 | ? | NA |
datadir <- datadir[md$sample_name]
print(all(names(datadir)==md$sample_name))
datadir[1] TRUE- donor1_fat
- '../data/donor1_fat/count/sample_filtered_feature_bc_matrix'
- donor1_skin
- '../data/donor1_skin/count/sample_filtered_feature_bc_matrix'
- donor2_fat
- '../data/donor2_fat/count/sample_filtered_feature_bc_matrix'
- donor3_fat
- '../data/donor3_fat/count/sample_filtered_feature_bc_matrix'
- donor3_skin
- '../data/donor3_skin/count/sample_filtered_feature_bc_matrix'
- donor4_fat
- '../data/donor4_fat/count/sample_filtered_feature_bc_matrix'
- donor4_skin
- '../data/donor4_skin/count/sample_filtered_feature_bc_matrix'
- donor5_recovery
- '../data/donor5_recovery/filtered_feature_bc_matrix'
- donor6_fat
- '../data/donor6_fat/count/sample_filtered_feature_bc_matrix'
- donor6_skin
- '../data/donor6_skin/count/sample_filtered_feature_bc_matrix'
- donor7_fat
- '../data/donor7_fat/count/sample_filtered_feature_bc_matrix'
- donor7_skin
- '../data/donor7_skin/count/sample_filtered_feature_bc_matrix'
fs <- split(datadir, md$sequencing_run)
fs <- lapply(fs, function(u) {
rc <- DropletUtils::read10xCounts(u)
nr <- nrow(rc)
if(any(grep("Multiplexing", rowData(rc)$Type))) {
rc <- rc[rowData(rc)$Type != "Multiplexing Capture",]
removed <- "barcodes"
}
else {
rc <- rc[-grep("NA.[1-6]", rownames(rc)),]
removed <- "features"
}
cat(sprintf(
"%s: removing %d %s; %d features left.\n", paste0(names(u), collapse=","),
nr-nrow(rc), removed, nrow(rc)
))
return(rc)
})
fsdonor1_fat,donor1_skin,donor2_fat: removing 3 barcodes; 21134 features left.
donor3_fat,donor3_skin,donor4_fat,donor4_skin: removing 4 barcodes; 21134 features left.
donor6_fat,donor6_skin,donor7_fat,donor7_skin: removing 6 barcodes; 21134 features left.
donor5_recovery: removing 6 features; 21134 features left.$`1`
class: SingleCellExperiment
dim: 21134 6891
metadata(1): Samples
assays(1): counts
rownames(21134): ENSG00000186092 ENSG00000284733 ... ENSG00000278817
ENSG00000277196
rowData names(3): ID Symbol Type
colnames: NULL
colData names(2): Sample Barcode
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):
$`2`
class: SingleCellExperiment
dim: 21134 2759
metadata(1): Samples
assays(1): counts
rownames(21134): ENSG00000186092 ENSG00000284733 ... ENSG00000278817
ENSG00000277196
rowData names(3): ID Symbol Type
colnames: NULL
colData names(2): Sample Barcode
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):
$`3`
class: SingleCellExperiment
dim: 21134 8336
metadata(1): Samples
assays(1): counts
rownames(21134): ENSG00000186092 ENSG00000284733 ... ENSG00000278817
ENSG00000277196
rowData names(3): ID Symbol Type
colnames: NULL
colData names(2): Sample Barcode
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):
$`4`
class: SingleCellExperiment
dim: 21134 4889
metadata(1): Samples
assays(1): counts
rownames(21134): ENSG00000186092 ENSG00000284733 ... ENSG00000278817
ENSG00000277196
rowData names(3): ID Symbol Type
colnames: NULL
colData names(2): Sample Barcode
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):sce <- SingleCellExperiment(
assays = SimpleList(
counts = do.call("cbind", lapply(fs, counts))
),
rowData = rowData(fs[[1]]),
colData = do.call("rbind", lapply(fs, colData))
)
table(sce$Sample)
donor1_fat donor1_skin donor2_fat donor3_fat donor3_skin
162 3861 2868 1133 620
donor4_fat donor4_skin donor5_recovery donor6_fat donor6_skin
131 875 4889 2001 1715
donor7_fat donor7_skin
1713 2907 colData(sce) <- colData(sce) %>% as.data.frame %>%
left_join(md %>% select(-keep,-notes), by = c("Sample"="sample_name")) %>% DataFrame
colData(sce)DataFrame with 22875 rows and 7 columns
Sample Barcode donor tissue tissue_origin
<character> <character> <numeric> <character> <character>
1 donor1_fat AAAGAACCACATACGT-1 1 fat abdomen
2 donor1_fat AAAGGTAGTCCGGTCA-1 1 fat abdomen
3 donor1_fat AAAGGTATCTCTCTTC-1 1 fat abdomen
4 donor1_fat AACCACACAGTGGCTC-1 1 fat abdomen
5 donor1_fat AACCCAAGTATGGGAC-1 1 fat abdomen
... ... ... ... ... ...
22871 donor5_recovery TTTGGTTTCGACCAAT-1 5 mixed abdomen
22872 donor5_recovery TTTGTTGGTCCTACGG-1 5 mixed abdomen
22873 donor5_recovery TTTGTTGGTCGCAGTC-1 5 mixed abdomen
22874 donor5_recovery TTTGTTGGTGGCGTAA-1 5 mixed abdomen
22875 donor5_recovery TTTGTTGTCGGCATTA-1 5 mixed abdomen
sequencing_run sex
<numeric> <character>
1 1 F
2 1 F
3 1 F
4 1 F
5 1 F
... ... ...
22871 4 ?
22872 4 ?
22873 4 ?
22874 4 ?
22875 4 ?colnames(sce) <- paste0(sce$Sample, ".", sce$Barcode)
rownames(sce) <- paste0(rowData(sce)$ID, ".", rowData(sce)$Symbol)
sceclass: SingleCellExperiment
dim: 21134 22875
metadata(0):
assays(1): counts
rownames(21134): ENSG00000186092.OR4F5 ENSG00000284733.OR4F29 ...
ENSG00000278817.ENSG00000278817 ENSG00000277196.ENSG00000277196
rowData names(3): ID Symbol Type
colnames(22875): donor1_fat.AAAGAACCACATACGT-1
donor1_fat.AAAGGTAGTCCGGTCA-1 ... donor5_recovery.TTTGTTGGTGGCGTAA-1
donor5_recovery.TTTGTTGTCGGCATTA-1
colData names(7): Sample Barcode ... sequencing_run sex
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):# very basic filter
sce <- sce[rowSums(counts(sce))>0]
sceclass: SingleCellExperiment
dim: 18664 22875
metadata(0):
assays(1): counts
rownames(18664): ENSG00000187634.SAMD11 ENSG00000188976.NOC2L ...
ENSG00000278817.ENSG00000278817 ENSG00000277196.ENSG00000277196
rowData names(3): ID Symbol Type
colnames(22875): donor1_fat.AAAGAACCACATACGT-1
donor1_fat.AAAGGTAGTCCGGTCA-1 ... donor5_recovery.TTTGTTGGTGGCGTAA-1
donor5_recovery.TTTGTTGTCGGCATTA-1
colData names(7): Sample Barcode ... sequencing_run sex
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):saveRDS(sce, file="../output/0.0-sce-unfiltered.rds")is.mito <- grepl("\\.mt-", rownames(sce), ignore.case = TRUE)
rownames(sce)[is.mito]
sum(is.mito)- 'ENSG00000210049.MT-TF'
- 'ENSG00000211459.MT-RNR1'
- 'ENSG00000210077.MT-TV'
- 'ENSG00000210082.MT-RNR2'
- 'ENSG00000209082.MT-TL1'
- 'ENSG00000198888.MT-ND1'
- 'ENSG00000210100.MT-TI'
- 'ENSG00000210107.MT-TQ'
- 'ENSG00000210112.MT-TM'
- 'ENSG00000198763.MT-ND2'
- 'ENSG00000210117.MT-TW'
- 'ENSG00000210127.MT-TA'
- 'ENSG00000210135.MT-TN'
- 'ENSG00000210140.MT-TC'
- 'ENSG00000210144.MT-TY'
- 'ENSG00000198804.MT-CO1'
- 'ENSG00000210151.MT-TS1'
- 'ENSG00000210154.MT-TD'
- 'ENSG00000198712.MT-CO2'
- 'ENSG00000210156.MT-TK'
- 'ENSG00000228253.MT-ATP8'
- 'ENSG00000198899.MT-ATP6'
- 'ENSG00000198938.MT-CO3'
- 'ENSG00000210164.MT-TG'
- 'ENSG00000198840.MT-ND3'
- 'ENSG00000210174.MT-TR'
- 'ENSG00000212907.MT-ND4L'
- 'ENSG00000198886.MT-ND4'
- 'ENSG00000210176.MT-TH'
- 'ENSG00000210184.MT-TS2'
- 'ENSG00000210191.MT-TL2'
- 'ENSG00000198786.MT-ND5'
- 'ENSG00000198695.MT-ND6'
- 'ENSG00000210194.MT-TE'
- 'ENSG00000198727.MT-CYB'
- 'ENSG00000210195.MT-TT'
- 'ENSG00000210196.MT-TP'
37
sce <- addPerCellQC(sce, subsets=list(Mito=is.mito), percent_top=c(50,100,200,500))
sce <- sce[!is.mito]
head(colData(sce),3)DataFrame with 3 rows and 17 columns
Sample Barcode donor
<character> <character> <numeric>
donor1_fat.AAAGAACCACATACGT-1 donor1_fat AAAGAACCACATACGT-1 1
donor1_fat.AAAGGTAGTCCGGTCA-1 donor1_fat AAAGGTAGTCCGGTCA-1 1
donor1_fat.AAAGGTATCTCTCTTC-1 donor1_fat AAAGGTATCTCTCTTC-1 1
tissue tissue_origin sequencing_run
<character> <character> <numeric>
donor1_fat.AAAGAACCACATACGT-1 fat abdomen 1
donor1_fat.AAAGGTAGTCCGGTCA-1 fat abdomen 1
donor1_fat.AAAGGTATCTCTCTTC-1 fat abdomen 1
sex sum detected percent.top_50
<character> <numeric> <integer> <numeric>
donor1_fat.AAAGAACCACATACGT-1 F 4276 1721 33.8868
donor1_fat.AAAGGTAGTCCGGTCA-1 F 3023 1084 39.6626
donor1_fat.AAAGGTATCTCTCTTC-1 F 3892 1422 34.1213
percent.top_100 percent.top_200 percent.top_500
<numeric> <numeric> <numeric>
donor1_fat.AAAGAACCACATACGT-1 42.7736 52.9935 69.3171
donor1_fat.AAAGGTAGTCCGGTCA-1 52.9937 65.0017 80.6814
donor1_fat.AAAGGTATCTCTCTTC-1 46.5570 58.3505 75.9250
subsets_Mito_sum subsets_Mito_detected
<numeric> <integer>
donor1_fat.AAAGAACCACATACGT-1 752 16
donor1_fat.AAAGGTAGTCCGGTCA-1 52 12
donor1_fat.AAAGGTATCTCTCTTC-1 48 12
subsets_Mito_percent total
<numeric> <numeric>
donor1_fat.AAAGAACCACATACGT-1 17.58653 4276
donor1_fat.AAAGGTAGTCCGGTCA-1 1.72015 3023
donor1_fat.AAAGGTATCTCTCTTC-1 1.23330 3892cd_df <- colData(sce) %>% as.data.frame
cd_df| Sample | Barcode | donor | tissue | tissue_origin | sequencing_run | sex | sum | detected | percent.top_50 | percent.top_100 | percent.top_200 | percent.top_500 | subsets_Mito_sum | subsets_Mito_detected | subsets_Mito_percent | total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <chr> | <dbl> | <chr> | <chr> | <dbl> | <chr> | <dbl> | <int> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <int> | <dbl> | <dbl> | |
| donor1_fat.AAAGAACCACATACGT-1 | donor1_fat | AAAGAACCACATACGT-1 | 1 | fat | abdomen | 1 | F | 4276 | 1721 | 33.88681 | 42.77362 | 52.99345 | 69.31712 | 752 | 16 | 17.5865295 | 4276 |
| donor1_fat.AAAGGTAGTCCGGTCA-1 | donor1_fat | AAAGGTAGTCCGGTCA-1 | 1 | fat | abdomen | 1 | F | 3023 | 1084 | 39.66259 | 52.99371 | 65.00165 | 80.68144 | 52 | 12 | 1.7201456 | 3023 |
| donor1_fat.AAAGGTATCTCTCTTC-1 | donor1_fat | AAAGGTATCTCTCTTC-1 | 1 | fat | abdomen | 1 | F | 3892 | 1422 | 34.12127 | 46.55704 | 58.35046 | 75.92497 | 48 | 12 | 1.2332991 | 3892 |
| donor1_fat.AACCACACAGTGGCTC-1 | donor1_fat | AACCACACAGTGGCTC-1 | 1 | fat | abdomen | 1 | F | 7722 | 2633 | 26.89718 | 38.12484 | 48.67910 | 64.12846 | 103 | 13 | 1.3338513 | 7722 |
| donor1_fat.AACCCAAGTATGGGAC-1 | donor1_fat | AACCCAAGTATGGGAC-1 | 1 | fat | abdomen | 1 | F | 636 | 178 | 79.87421 | 87.73585 | 100.00000 | 100.00000 | 445 | 15 | 69.9685535 | 636 |
| donor1_fat.AACGGGATCTGCTGAA-1 | donor1_fat | AACGGGATCTGCTGAA-1 | 1 | fat | abdomen | 1 | F | 3256 | 1284 | 34.33661 | 46.80590 | 59.06020 | 75.92138 | 123 | 11 | 3.7776413 | 3256 |
| donor1_fat.AAGAACATCTGTGCAA-1 | donor1_fat | AAGAACATCTGTGCAA-1 | 1 | fat | abdomen | 1 | F | 6777 | 1989 | 32.86115 | 47.38085 | 60.01180 | 74.07407 | 224 | 14 | 3.3052973 | 6777 |
| donor1_fat.AAGCATCTCGATCCCT-1 | donor1_fat | AAGCATCTCGATCCCT-1 | 1 | fat | abdomen | 1 | F | 1746 | 839 | 34.07789 | 47.53723 | 61.45475 | 80.58419 | 63 | 11 | 3.6082474 | 1746 |
| donor1_fat.AAGCGAGAGTATCCTG-1 | donor1_fat | AAGCGAGAGTATCCTG-1 | 1 | fat | abdomen | 1 | F | 6750 | 2393 | 24.08889 | 35.12593 | 46.69630 | 63.71852 | 277 | 18 | 4.1037037 | 6750 |
| donor1_fat.AAGTTCGAGGCAGGTT-1 | donor1_fat | AAGTTCGAGGCAGGTT-1 | 1 | fat | abdomen | 1 | F | 5707 | 2041 | 31.20729 | 43.42036 | 54.17908 | 68.91537 | 188 | 12 | 3.2942001 | 5707 |
| donor1_fat.AATCGACCAATTGCAC-1 | donor1_fat | AATCGACCAATTGCAC-1 | 1 | fat | abdomen | 1 | F | 3401 | 1534 | 26.60982 | 37.37136 | 49.51485 | 68.92091 | 107 | 15 | 3.1461335 | 3401 |
| donor1_fat.ACAAAGACAATGACCT-1 | donor1_fat | ACAAAGACAATGACCT-1 | 1 | fat | abdomen | 1 | F | 5851 | 1802 | 32.49017 | 45.70159 | 58.43446 | 74.67100 | 106 | 16 | 1.8116561 | 5851 |
| donor1_fat.ACAAAGACACTAACGT-1 | donor1_fat | ACAAAGACACTAACGT-1 | 1 | fat | abdomen | 1 | F | 3602 | 1004 | 43.80900 | 59.35591 | 72.01555 | 86.00777 | 60 | 11 | 1.6657413 | 3602 |
| donor1_fat.ACAACCAAGGGCTTCC-1 | donor1_fat | ACAACCAAGGGCTTCC-1 | 1 | fat | abdomen | 1 | F | 10238 | 3273 | 22.24067 | 33.03380 | 44.50088 | 59.89451 | 168 | 12 | 1.6409455 | 10238 |
| donor1_fat.ACAGGGACACACTTAG-1 | donor1_fat | ACAGGGACACACTTAG-1 | 1 | fat | abdomen | 1 | F | 606 | 63 | 97.85479 | 100.00000 | 100.00000 | 100.00000 | 556 | 15 | 91.7491749 | 606 |
| donor1_fat.ACAGGGATCTCATAGG-1 | donor1_fat | ACAGGGATCTCATAGG-1 | 1 | fat | abdomen | 1 | F | 5210 | 1884 | 27.96545 | 40.07678 | 53.03263 | 69.17466 | 105 | 13 | 2.0153551 | 5210 |
| donor1_fat.ACATTTCCACTCCACT-1 | donor1_fat | ACATTTCCACTCCACT-1 | 1 | fat | abdomen | 1 | F | 3606 | 1305 | 36.30061 | 49.16805 | 61.53633 | 77.67610 | 33 | 11 | 0.9151414 | 3606 |
| donor1_fat.ACATTTCTCGATGGAG-1 | donor1_fat | ACATTTCTCGATGGAG-1 | 1 | fat | abdomen | 1 | F | 3853 | 1491 | 34.07734 | 47.31378 | 58.44796 | 74.27978 | 68 | 13 | 1.7648586 | 3853 |
| donor1_fat.ACCCTTGTCACCATAG-1 | donor1_fat | ACCCTTGTCACCATAG-1 | 1 | fat | abdomen | 1 | F | 4627 | 1604 | 34.92544 | 48.32505 | 59.39053 | 74.82170 | 84 | 12 | 1.8154312 | 4627 |
| donor1_fat.ACCTACCAGACCTCCG-1 | donor1_fat | ACCTACCAGACCTCCG-1 | 1 | fat | abdomen | 1 | F | 1732 | 651 | 46.76674 | 60.56582 | 73.78753 | 91.28176 | 41 | 12 | 2.3672055 | 1732 |
| donor1_fat.ACGGAAGCACACCTAA-1 | donor1_fat | ACGGAAGCACACCTAA-1 | 1 | fat | abdomen | 1 | F | 5069 | 2279 | 17.65634 | 25.42908 | 36.41744 | 56.77648 | 369 | 14 | 7.2795423 | 5069 |
| donor1_fat.ACGTAACAGTCTAGCT-1 | donor1_fat | ACGTAACAGTCTAGCT-1 | 1 | fat | abdomen | 1 | F | 1188 | 491 | 60.43771 | 67.08754 | 75.50505 | 100.00000 | 620 | 18 | 52.1885522 | 1188 |
| donor1_fat.ACGTAGTTCCCGAAAT-1 | donor1_fat | ACGTAGTTCCCGAAAT-1 | 1 | fat | abdomen | 1 | F | 3897 | 1246 | 36.23300 | 49.73056 | 62.63793 | 80.70310 | 39 | 11 | 1.0007698 | 3897 |
| donor1_fat.ACGTTCCAGATGAATC-1 | donor1_fat | ACGTTCCAGATGAATC-1 | 1 | fat | abdomen | 1 | F | 1884 | 1136 | 28.60934 | 36.57113 | 47.29299 | 66.24204 | 280 | 13 | 14.8619958 | 1884 |
| donor1_fat.ACTCTCGAGCCGTTGC-1 | donor1_fat | ACTCTCGAGCCGTTGC-1 | 1 | fat | abdomen | 1 | F | 1220 | 611 | 41.96721 | 53.77049 | 66.31148 | 90.90164 | 154 | 15 | 12.6229508 | 1220 |
| donor1_fat.ACTTCCGTCAGGAACG-1 | donor1_fat | ACTTCCGTCAGGAACG-1 | 1 | fat | abdomen | 1 | F | 4898 | 2364 | 17.29277 | 24.37730 | 34.29971 | 53.14414 | 367 | 16 | 7.4928542 | 4898 |
| donor1_fat.AGACACTCAGGCGATA-1 | donor1_fat | AGACACTCAGGCGATA-1 | 1 | fat | abdomen | 1 | F | 4601 | 1518 | 33.51445 | 46.70724 | 59.70441 | 76.26603 | 171 | 12 | 3.7165834 | 4601 |
| donor1_fat.AGACCATTCAAAGGTA-1 | donor1_fat | AGACCATTCAAAGGTA-1 | 1 | fat | abdomen | 1 | F | 7030 | 2375 | 27.28307 | 38.50640 | 50.54054 | 66.28734 | 27 | 10 | 0.3840683 | 7030 |
| donor1_fat.AGAGAGCCAAGCCCAC-1 | donor1_fat | AGAGAGCCAAGCCCAC-1 | 1 | fat | abdomen | 1 | F | 3437 | 1047 | 43.26447 | 57.81205 | 69.53739 | 84.08496 | 85 | 12 | 2.4730870 | 3437 |
| donor1_fat.AGAGCCCAGTCTAGCT-1 | donor1_fat | AGAGCCCAGTCTAGCT-1 | 1 | fat | abdomen | 1 | F | 4781 | 1680 | 33.44489 | 46.37105 | 58.12592 | 73.41560 | 110 | 14 | 2.3007739 | 4781 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| donor5_recovery.TTTCGATAGTGGAATT-1 | donor5_recovery | TTTCGATAGTGGAATT-1 | 5 | mixed | abdomen | 4 | ? | 3048 | 1575 | 21.42388 | 31.33202 | 42.84777 | 64.20604 | 139 | 16 | 4.5603675 | 3048 |
| donor5_recovery.TTTCGATGTGCTGATT-1 | donor5_recovery | TTTCGATGTGCTGATT-1 | 5 | mixed | abdomen | 4 | ? | 2238 | 1400 | 17.20286 | 25.24576 | 36.90795 | 59.78552 | 57 | 15 | 2.5469169 | 2238 |
| donor5_recovery.TTTGACTAGATGGGCT-1 | donor5_recovery | TTTGACTAGATGGGCT-1 | 5 | mixed | abdomen | 4 | ? | 2448 | 1208 | 27.69608 | 39.95098 | 53.10458 | 71.07843 | 95 | 16 | 3.8807190 | 2448 |
| donor5_recovery.TTTGACTAGTGTTCAC-1 | donor5_recovery | TTTGACTAGTGTTCAC-1 | 5 | mixed | abdomen | 4 | ? | 6499 | 2564 | 24.23450 | 34.06678 | 44.72996 | 60.70165 | 78 | 13 | 1.2001846 | 6499 |
| donor5_recovery.TTTGACTGTCCAAATC-1 | donor5_recovery | TTTGACTGTCCAAATC-1 | 5 | mixed | abdomen | 4 | ? | 958 | 673 | 24.32150 | 35.80376 | 50.62630 | 81.94154 | 8 | 4 | 0.8350731 | 958 |
| donor5_recovery.TTTGACTGTGATTCTG-1 | donor5_recovery | TTTGACTGTGATTCTG-1 | 5 | mixed | abdomen | 4 | ? | 1568 | 955 | 26.97704 | 37.75510 | 50.70153 | 70.98214 | 71 | 14 | 4.5280612 | 1568 |
| donor5_recovery.TTTGACTGTTGTCTAG-1 | donor5_recovery | TTTGACTGTTGTCTAG-1 | 5 | mixed | abdomen | 4 | ? | 936 | 593 | 29.70085 | 42.30769 | 58.01282 | 90.06410 | 38 | 11 | 4.0598291 | 936 |
| donor5_recovery.TTTGATCAGATGACCG-1 | donor5_recovery | TTTGATCAGATGACCG-1 | 5 | mixed | abdomen | 4 | ? | 956 | 596 | 30.75314 | 43.61925 | 58.57741 | 89.95816 | 25 | 8 | 2.6150628 | 956 |
| donor5_recovery.TTTGATCAGCAATAAC-1 | donor5_recovery | TTTGATCAGCAATAAC-1 | 5 | mixed | abdomen | 4 | ? | 2148 | 939 | 34.35754 | 48.51024 | 61.17318 | 79.56238 | 71 | 14 | 3.3054004 | 2148 |
| donor5_recovery.TTTGATCAGTTACTCG-1 | donor5_recovery | TTTGATCAGTTACTCG-1 | 5 | mixed | abdomen | 4 | ? | 3145 | 1486 | 21.90779 | 32.84579 | 46.45469 | 67.59936 | 66 | 15 | 2.0985692 | 3145 |
| donor5_recovery.TTTGATCGTTGGGCCT-1 | donor5_recovery | TTTGATCGTTGGGCCT-1 | 5 | mixed | abdomen | 4 | ? | 2897 | 1316 | 30.54884 | 42.18157 | 54.57370 | 71.83293 | 182 | 17 | 6.2823611 | 2897 |
| donor5_recovery.TTTGATCTCAAAGGTA-1 | donor5_recovery | TTTGATCTCAAAGGTA-1 | 5 | mixed | abdomen | 4 | ? | 2155 | 1136 | 26.07889 | 36.98376 | 50.06961 | 70.48724 | 98 | 14 | 4.5475638 | 2155 |
| donor5_recovery.TTTGGAGAGTGCAAAT-1 | donor5_recovery | TTTGGAGAGTGCAAAT-1 | 5 | mixed | abdomen | 4 | ? | 1054 | 601 | 34.91461 | 48.10247 | 61.95446 | 90.41746 | 43 | 14 | 4.0796964 | 1054 |
| donor5_recovery.TTTGGAGCATAGGTAA-1 | donor5_recovery | TTTGGAGCATAGGTAA-1 | 5 | mixed | abdomen | 4 | ? | 1846 | 1050 | 23.34778 | 33.91116 | 47.50813 | 70.20585 | 89 | 14 | 4.8212351 | 1846 |
| donor5_recovery.TTTGGAGGTCAGTTTG-1 | donor5_recovery | TTTGGAGGTCAGTTTG-1 | 5 | mixed | abdomen | 4 | ? | 1222 | 769 | 26.67758 | 38.21604 | 53.43699 | 77.98691 | 51 | 13 | 4.1734861 | 1222 |
| donor5_recovery.TTTGGAGGTCCAAGAG-1 | donor5_recovery | TTTGGAGGTCCAAGAG-1 | 5 | mixed | abdomen | 4 | ? | 1829 | 1058 | 27.28267 | 37.88956 | 49.64461 | 69.49153 | 28 | 12 | 1.5308912 | 1829 |
| donor5_recovery.TTTGGAGGTCGACTTA-1 | donor5_recovery | TTTGGAGGTCGACTTA-1 | 5 | mixed | abdomen | 4 | ? | 2389 | 1430 | 17.91545 | 26.28715 | 38.21683 | 61.07158 | 154 | 21 | 6.4462118 | 2389 |
| donor5_recovery.TTTGGAGGTTGAGAGC-1 | donor5_recovery | TTTGGAGGTTGAGAGC-1 | 5 | mixed | abdomen | 4 | ? | 1618 | 993 | 21.69345 | 32.26205 | 46.66255 | 69.53028 | 38 | 11 | 2.3485785 | 1618 |
| donor5_recovery.TTTGGAGTCCAAGAGG-1 | donor5_recovery | TTTGGAGTCCAAGAGG-1 | 5 | mixed | abdomen | 4 | ? | 1262 | 830 | 23.85103 | 34.62758 | 50.07924 | 73.85103 | 69 | 14 | 5.4675119 | 1262 |
| donor5_recovery.TTTGGAGTCTCAACCC-1 | donor5_recovery | TTTGGAGTCTCAACCC-1 | 5 | mixed | abdomen | 4 | ? | 1135 | 717 | 29.51542 | 40.79295 | 54.44934 | 80.88106 | 18 | 10 | 1.5859031 | 1135 |
| donor5_recovery.TTTGGTTAGCCTCTTC-1 | donor5_recovery | TTTGGTTAGCCTCTTC-1 | 5 | mixed | abdomen | 4 | ? | 763 | 584 | 24.90170 | 36.56619 | 49.67235 | 88.99083 | 24 | 8 | 3.1454784 | 763 |
| donor5_recovery.TTTGGTTCACTCTAGA-1 | donor5_recovery | TTTGGTTCACTCTAGA-1 | 5 | mixed | abdomen | 4 | ? | 812 | 570 | 27.58621 | 40.14778 | 54.43350 | 91.37931 | 24 | 8 | 2.9556650 | 812 |
| donor5_recovery.TTTGGTTGTACGTGAG-1 | donor5_recovery | TTTGGTTGTACGTGAG-1 | 5 | mixed | abdomen | 4 | ? | 755 | 494 | 30.99338 | 44.90066 | 61.05960 | 100.00000 | 34 | 9 | 4.5033113 | 755 |
| donor5_recovery.TTTGGTTGTCCTACAA-1 | donor5_recovery | TTTGGTTGTCCTACAA-1 | 5 | mixed | abdomen | 4 | ? | 835 | 625 | 24.07186 | 36.04790 | 49.10180 | 85.02994 | 27 | 6 | 3.2335329 | 835 |
| donor5_recovery.TTTGGTTGTGACTCTA-1 | donor5_recovery | TTTGGTTGTGACTCTA-1 | 5 | mixed | abdomen | 4 | ? | 938 | 572 | 29.63753 | 43.39019 | 60.34115 | 92.32409 | 2 | 2 | 0.2132196 | 938 |
| donor5_recovery.TTTGGTTTCGACCAAT-1 | donor5_recovery | TTTGGTTTCGACCAAT-1 | 5 | mixed | abdomen | 4 | ? | 1328 | 670 | 34.33735 | 48.34337 | 64.30723 | 87.19880 | 67 | 10 | 5.0451807 | 1328 |
| donor5_recovery.TTTGTTGGTCCTACGG-1 | donor5_recovery | TTTGTTGGTCCTACGG-1 | 5 | mixed | abdomen | 4 | ? | 795 | 552 | 28.30189 | 41.63522 | 55.72327 | 93.45912 | 44 | 12 | 5.5345912 | 795 |
| donor5_recovery.TTTGTTGGTCGCAGTC-1 | donor5_recovery | TTTGTTGGTCGCAGTC-1 | 5 | mixed | abdomen | 4 | ? | 1872 | 1250 | 16.18590 | 24.73291 | 37.01923 | 59.93590 | 66 | 14 | 3.5256410 | 1872 |
| donor5_recovery.TTTGTTGGTGGCGTAA-1 | donor5_recovery | TTTGTTGGTGGCGTAA-1 | 5 | mixed | abdomen | 4 | ? | 1928 | 931 | 33.14315 | 46.21369 | 58.97303 | 77.64523 | 51 | 13 | 2.6452282 | 1928 |
| donor5_recovery.TTTGTTGTCGGCATTA-1 | donor5_recovery | TTTGTTGTCGGCATTA-1 | 5 | mixed | abdomen | 4 | ? | 2091 | 1206 | 21.95122 | 32.56815 | 45.28934 | 66.23625 | 53 | 14 | 2.5346724 | 2091 |
options(repr.plot.width=20, repr.plot.height=20)
ggplot(cd_df, aes(sum, detected)) +
geom_point(size=.5) +
scale_x_log10() +
scale_y_log10() +
geom_density_2d(contour_var = "ndensity") +
facet_wrap(~Sample)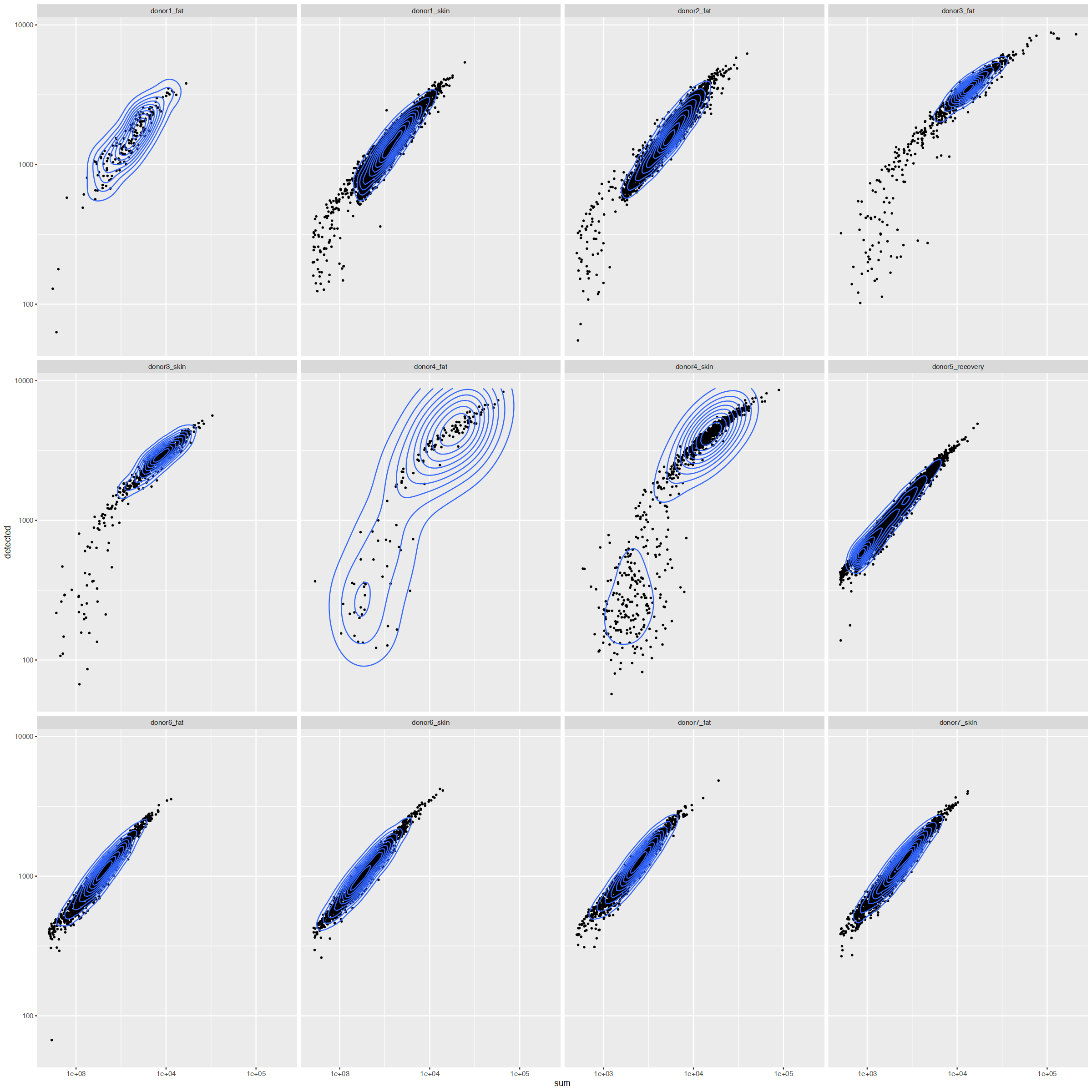
options(repr.plot.width=12, repr.plot.height=6)
ggplot(cd_df, aes(x=sum, color=Sample)) +
geom_density(size=4) +
scale_x_log10()Warning message:
“Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.”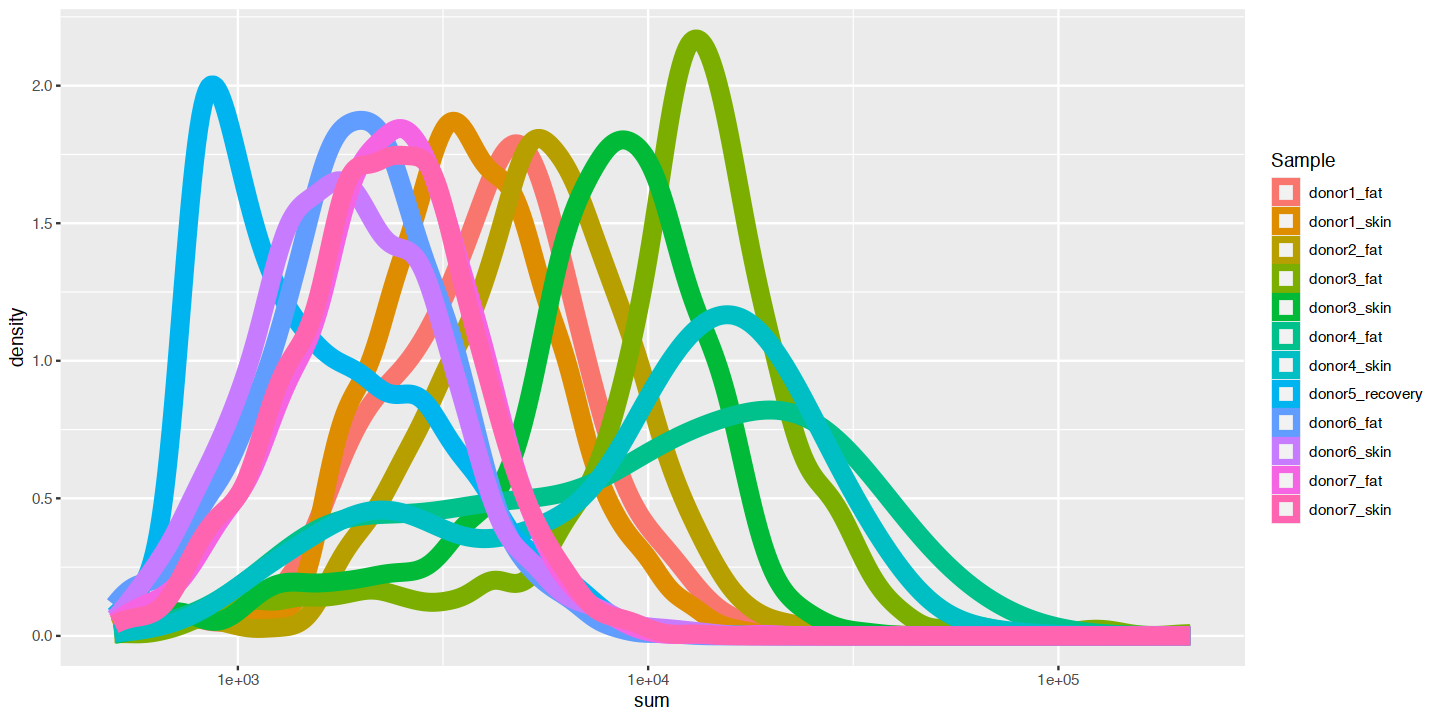
options(repr.plot.width=12, repr.plot.height=6)
ggplot(cd_df, aes(x=detected, color=Sample)) +
geom_density(linewidth=4) +
scale_x_log10()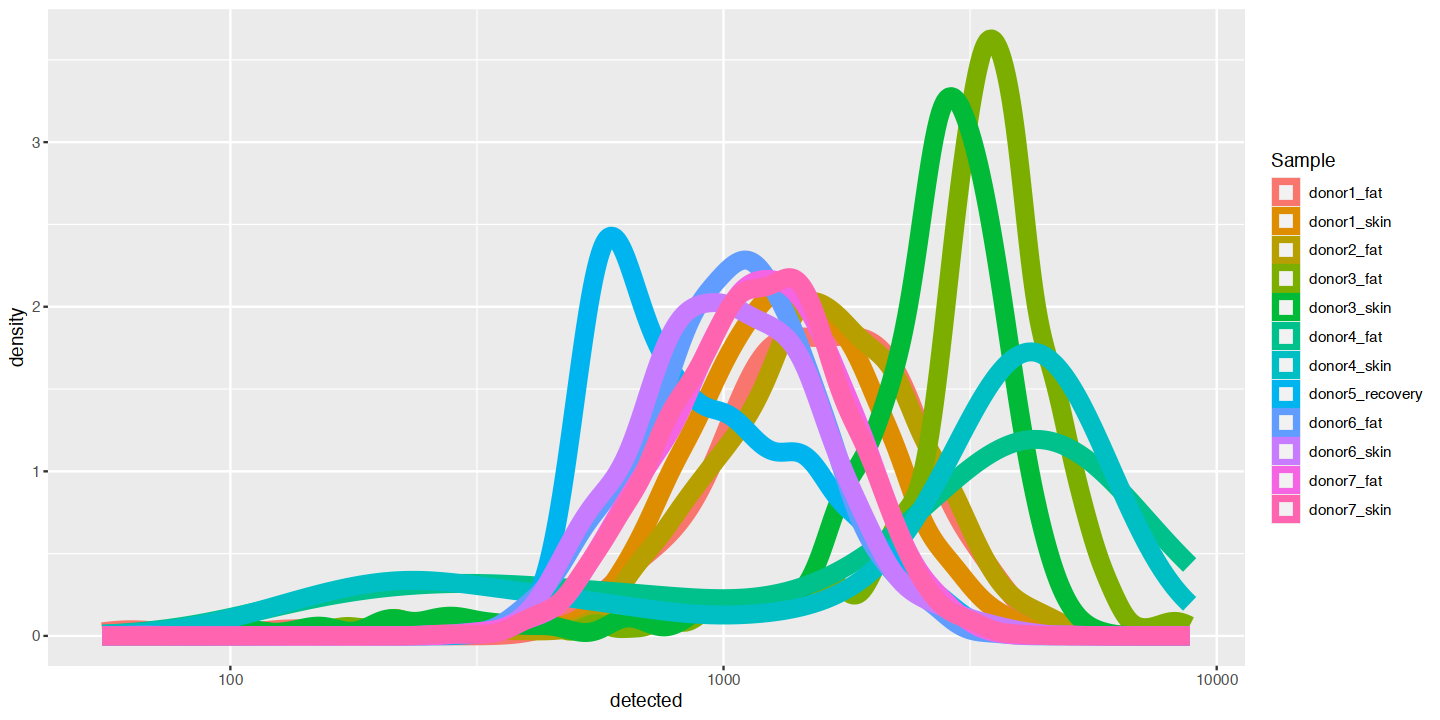
options(repr.plot.width=12, repr.plot.height=12)
ggplot(cd_df, aes(x=Sample, y=sum)) +
geom_violin() +
geom_jitter() +
scale_y_log10()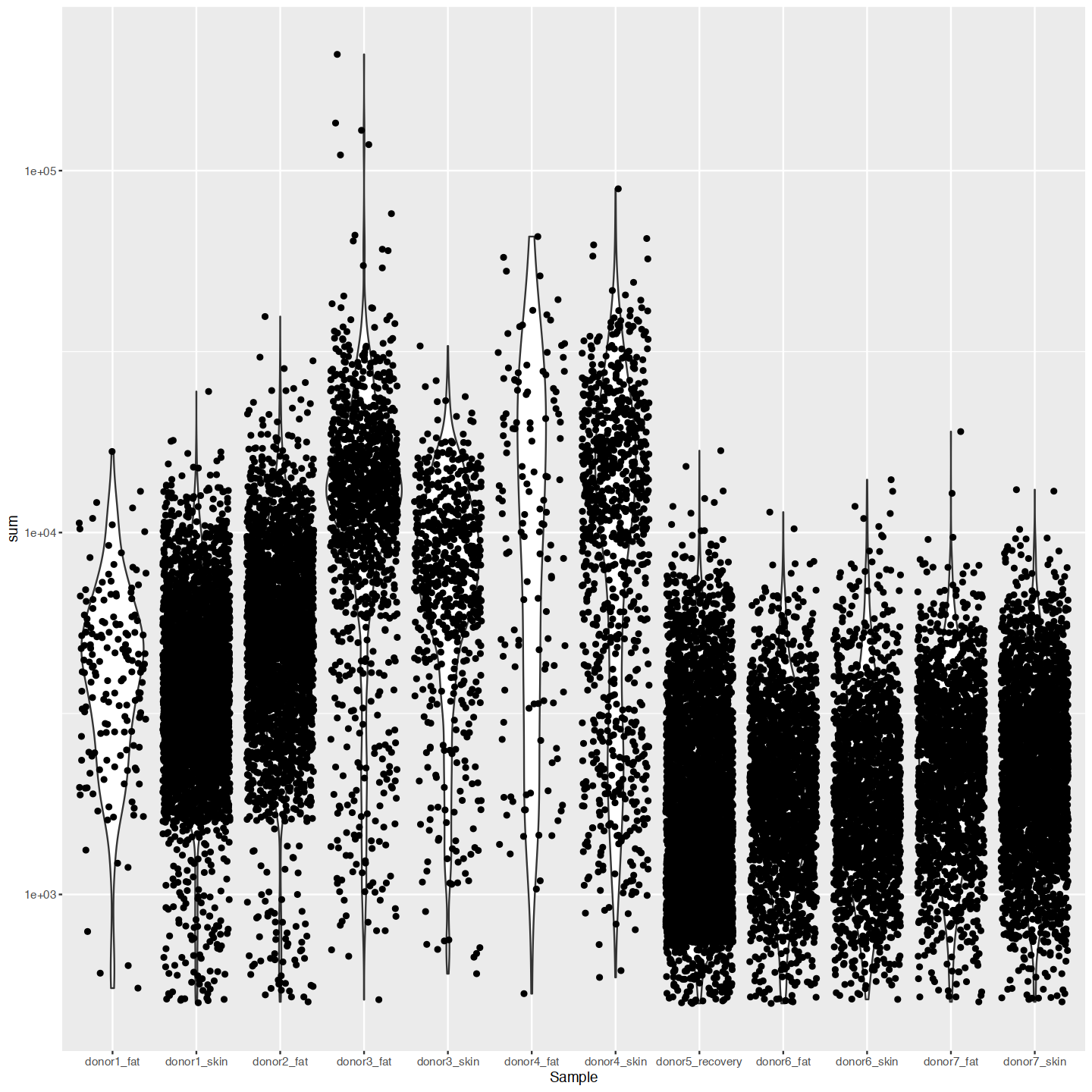
options(repr.plot.width=20, repr.plot.height=20)
ggplot(cd_df, aes(sum, percent.top_50)) +
geom_point(size=.5) +
scale_x_log10() +
geom_density_2d(contour_var = "ndensity") +
facet_wrap(~Sample)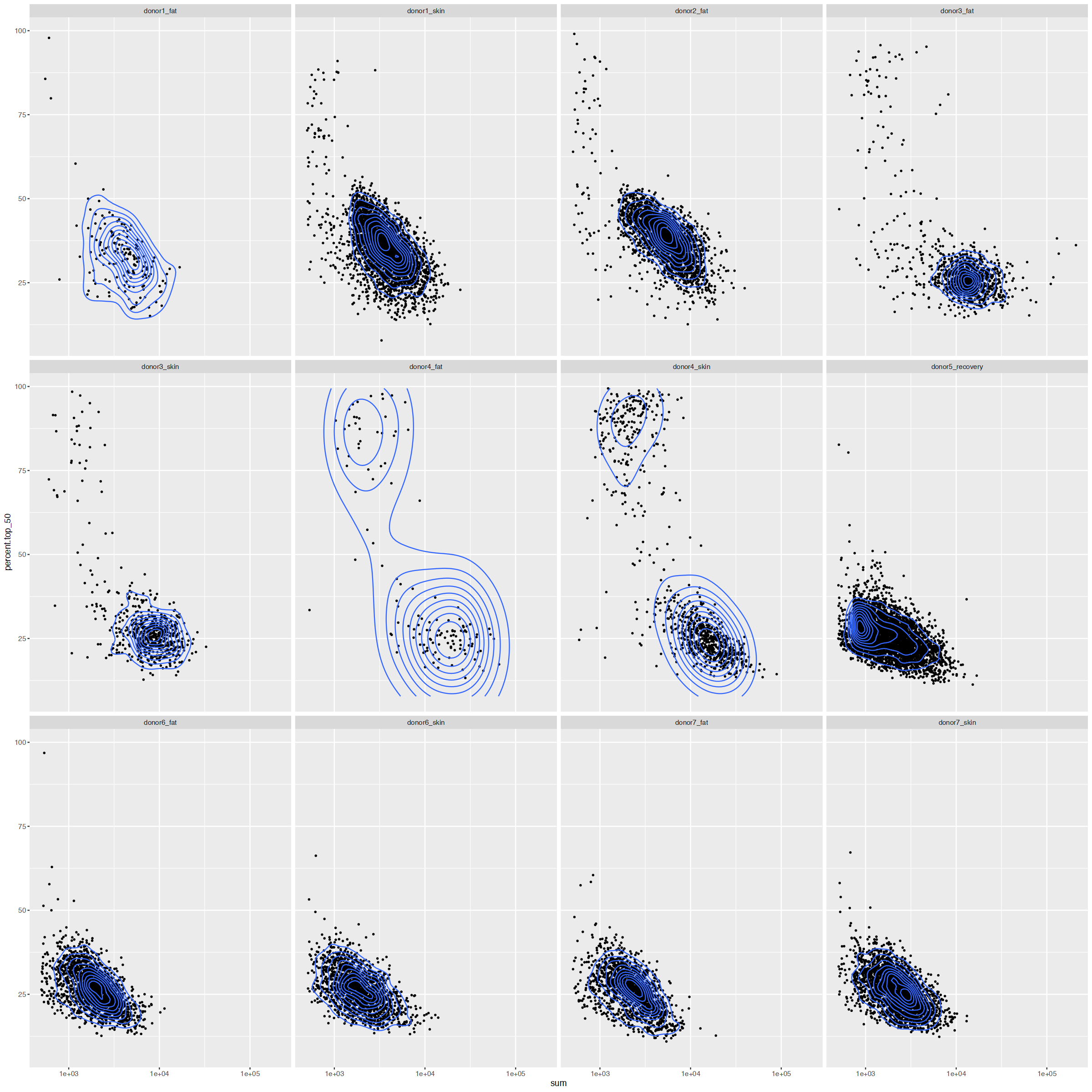
options(repr.plot.width=20, repr.plot.height=20)
ggplot(cd_df, aes(sum, subsets_Mito_percent)) +
geom_point(size=.5) +
scale_x_log10() +
scale_y_sqrt() +
geom_density_2d(contour_var = "ndensity") +
facet_wrap(~Sample)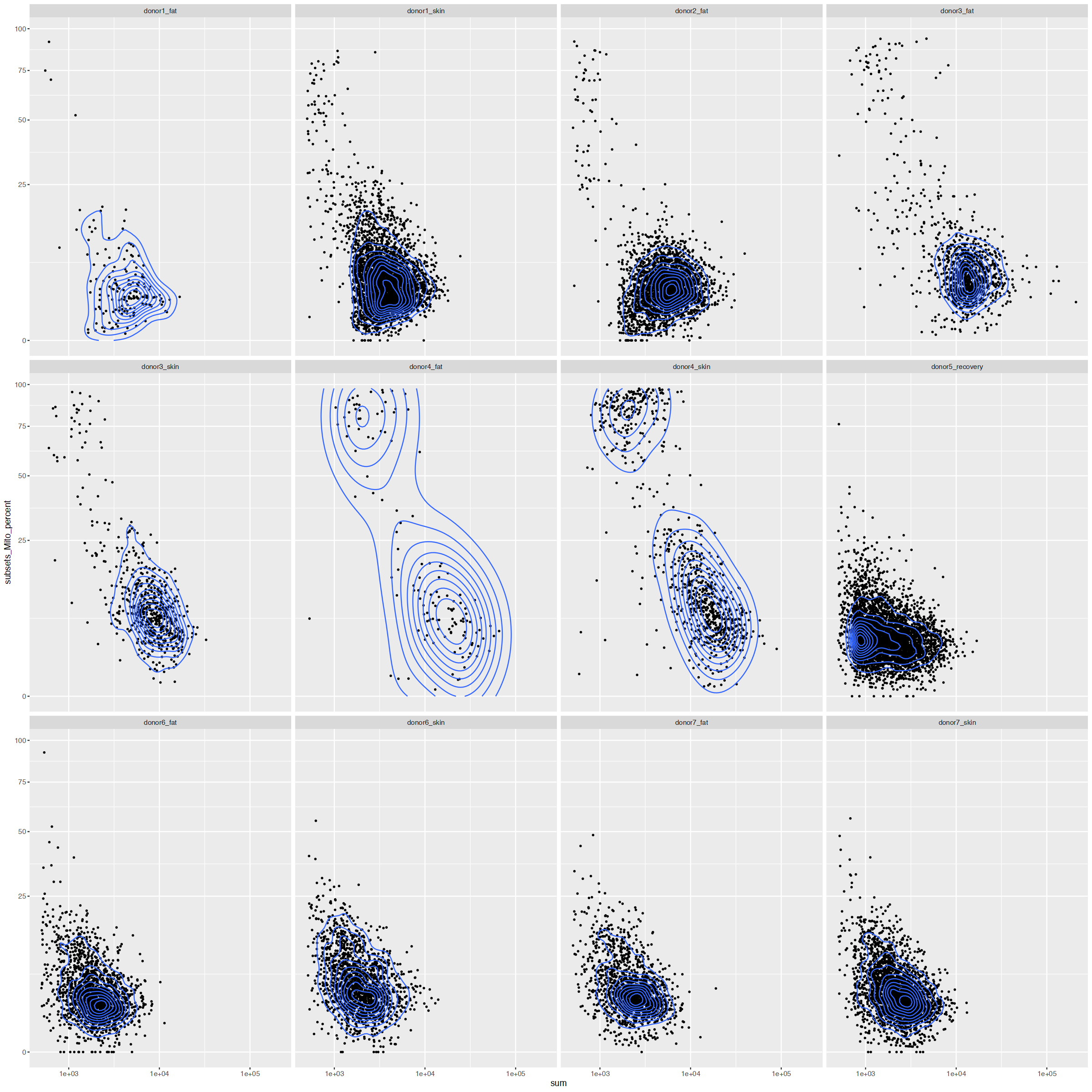
mito_cutoff <- 30
detected_cutoff <- 200with(cd_df, table(Sample, detected < detected_cutoff))
with(cd_df, table(Sample, subsets_Mito_percent > mito_cutoff))
with(cd_df, table(Sample, subsets_Mito_percent > mito_cutoff | detected < detected_cutoff))
Sample FALSE TRUE
donor1_fat 159 3
donor1_skin 3844 17
donor2_fat 2852 16
donor3_fat 1122 11
donor3_skin 611 9
donor4_fat 123 8
donor4_skin 810 65
donor5_recovery 4887 2
donor6_fat 2000 1
donor6_skin 1715 0
donor7_fat 1713 0
donor7_skin 2907 0
Sample FALSE TRUE
donor1_fat 158 4
donor1_skin 3804 57
donor2_fat 2823 45
donor3_fat 1063 70
donor3_skin 573 47
donor4_fat 89 42
donor4_skin 638 237
donor5_recovery 4880 9
donor6_fat 1994 7
donor6_skin 1710 5
donor7_fat 1708 5
donor7_skin 2898 9
Sample FALSE TRUE
donor1_fat 158 4
donor1_skin 3804 57
donor2_fat 2823 45
donor3_fat 1063 70
donor3_skin 573 47
donor4_fat 89 42
donor4_skin 638 237
donor5_recovery 4879 10
donor6_fat 1994 7
donor6_skin 1710 5
donor7_fat 1708 5
donor7_skin 2898 9sce <- sce[, (sce$subsets_Mito_percent < mito_cutoff) & (sce$detected > detected_cutoff)]
sceclass: SingleCellExperiment
dim: 18627 22337
metadata(0):
assays(1): counts
rownames(18627): ENSG00000187634.SAMD11 ENSG00000188976.NOC2L ...
ENSG00000278817.ENSG00000278817 ENSG00000277196.ENSG00000277196
rowData names(3): ID Symbol Type
colnames(22337): donor1_fat.AAAGAACCACATACGT-1
donor1_fat.AAAGGTAGTCCGGTCA-1 ... donor5_recovery.TTTGTTGGTGGCGTAA-1
donor5_recovery.TTTGTTGTCGGCATTA-1
colData names(17): Sample Barcode ... subsets_Mito_percent total
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):table(sce$tissue)
fat mixed skin
7835 4879 9623 cd_df <- colData(sce) %>% as.data.frame
cd_df <- cd_df %>% mutate(
outlier_sum=isOutlier(
sum, nmads=4, type="both", log=TRUE, batch=Sample
),
outlier_detected=isOutlier(
detected, nmads=4, type="both", log=TRUE, batch=Sample
)
)
cd_df| Sample | Barcode | donor | tissue | tissue_origin | sequencing_run | sex | sum | detected | percent.top_50 | percent.top_100 | percent.top_200 | percent.top_500 | subsets_Mito_sum | subsets_Mito_detected | subsets_Mito_percent | total | outlier_sum | outlier_detected | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <chr> | <dbl> | <chr> | <chr> | <dbl> | <chr> | <dbl> | <int> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <int> | <dbl> | <dbl> | <otlr.flt> | <otlr.flt> | |
| donor1_fat.AAAGAACCACATACGT-1 | donor1_fat | AAAGAACCACATACGT-1 | 1 | fat | abdomen | 1 | F | 4276 | 1721 | 33.88681 | 42.77362 | 52.99345 | 69.31712 | 752 | 16 | 17.5865295 | 4276 | FALSE | FALSE |
| donor1_fat.AAAGGTAGTCCGGTCA-1 | donor1_fat | AAAGGTAGTCCGGTCA-1 | 1 | fat | abdomen | 1 | F | 3023 | 1084 | 39.66259 | 52.99371 | 65.00165 | 80.68144 | 52 | 12 | 1.7201456 | 3023 | FALSE | FALSE |
| donor1_fat.AAAGGTATCTCTCTTC-1 | donor1_fat | AAAGGTATCTCTCTTC-1 | 1 | fat | abdomen | 1 | F | 3892 | 1422 | 34.12127 | 46.55704 | 58.35046 | 75.92497 | 48 | 12 | 1.2332991 | 3892 | FALSE | FALSE |
| donor1_fat.AACCACACAGTGGCTC-1 | donor1_fat | AACCACACAGTGGCTC-1 | 1 | fat | abdomen | 1 | F | 7722 | 2633 | 26.89718 | 38.12484 | 48.67910 | 64.12846 | 103 | 13 | 1.3338513 | 7722 | FALSE | FALSE |
| donor1_fat.AACGGGATCTGCTGAA-1 | donor1_fat | AACGGGATCTGCTGAA-1 | 1 | fat | abdomen | 1 | F | 3256 | 1284 | 34.33661 | 46.80590 | 59.06020 | 75.92138 | 123 | 11 | 3.7776413 | 3256 | FALSE | FALSE |
| donor1_fat.AAGAACATCTGTGCAA-1 | donor1_fat | AAGAACATCTGTGCAA-1 | 1 | fat | abdomen | 1 | F | 6777 | 1989 | 32.86115 | 47.38085 | 60.01180 | 74.07407 | 224 | 14 | 3.3052973 | 6777 | FALSE | FALSE |
| donor1_fat.AAGCATCTCGATCCCT-1 | donor1_fat | AAGCATCTCGATCCCT-1 | 1 | fat | abdomen | 1 | F | 1746 | 839 | 34.07789 | 47.53723 | 61.45475 | 80.58419 | 63 | 11 | 3.6082474 | 1746 | FALSE | FALSE |
| donor1_fat.AAGCGAGAGTATCCTG-1 | donor1_fat | AAGCGAGAGTATCCTG-1 | 1 | fat | abdomen | 1 | F | 6750 | 2393 | 24.08889 | 35.12593 | 46.69630 | 63.71852 | 277 | 18 | 4.1037037 | 6750 | FALSE | FALSE |
| donor1_fat.AAGTTCGAGGCAGGTT-1 | donor1_fat | AAGTTCGAGGCAGGTT-1 | 1 | fat | abdomen | 1 | F | 5707 | 2041 | 31.20729 | 43.42036 | 54.17908 | 68.91537 | 188 | 12 | 3.2942001 | 5707 | FALSE | FALSE |
| donor1_fat.AATCGACCAATTGCAC-1 | donor1_fat | AATCGACCAATTGCAC-1 | 1 | fat | abdomen | 1 | F | 3401 | 1534 | 26.60982 | 37.37136 | 49.51485 | 68.92091 | 107 | 15 | 3.1461335 | 3401 | FALSE | FALSE |
| donor1_fat.ACAAAGACAATGACCT-1 | donor1_fat | ACAAAGACAATGACCT-1 | 1 | fat | abdomen | 1 | F | 5851 | 1802 | 32.49017 | 45.70159 | 58.43446 | 74.67100 | 106 | 16 | 1.8116561 | 5851 | FALSE | FALSE |
| donor1_fat.ACAAAGACACTAACGT-1 | donor1_fat | ACAAAGACACTAACGT-1 | 1 | fat | abdomen | 1 | F | 3602 | 1004 | 43.80900 | 59.35591 | 72.01555 | 86.00777 | 60 | 11 | 1.6657413 | 3602 | FALSE | FALSE |
| donor1_fat.ACAACCAAGGGCTTCC-1 | donor1_fat | ACAACCAAGGGCTTCC-1 | 1 | fat | abdomen | 1 | F | 10238 | 3273 | 22.24067 | 33.03380 | 44.50088 | 59.89451 | 168 | 12 | 1.6409455 | 10238 | FALSE | FALSE |
| donor1_fat.ACAGGGATCTCATAGG-1 | donor1_fat | ACAGGGATCTCATAGG-1 | 1 | fat | abdomen | 1 | F | 5210 | 1884 | 27.96545 | 40.07678 | 53.03263 | 69.17466 | 105 | 13 | 2.0153551 | 5210 | FALSE | FALSE |
| donor1_fat.ACATTTCCACTCCACT-1 | donor1_fat | ACATTTCCACTCCACT-1 | 1 | fat | abdomen | 1 | F | 3606 | 1305 | 36.30061 | 49.16805 | 61.53633 | 77.67610 | 33 | 11 | 0.9151414 | 3606 | FALSE | FALSE |
| donor1_fat.ACATTTCTCGATGGAG-1 | donor1_fat | ACATTTCTCGATGGAG-1 | 1 | fat | abdomen | 1 | F | 3853 | 1491 | 34.07734 | 47.31378 | 58.44796 | 74.27978 | 68 | 13 | 1.7648586 | 3853 | FALSE | FALSE |
| donor1_fat.ACCCTTGTCACCATAG-1 | donor1_fat | ACCCTTGTCACCATAG-1 | 1 | fat | abdomen | 1 | F | 4627 | 1604 | 34.92544 | 48.32505 | 59.39053 | 74.82170 | 84 | 12 | 1.8154312 | 4627 | FALSE | FALSE |
| donor1_fat.ACCTACCAGACCTCCG-1 | donor1_fat | ACCTACCAGACCTCCG-1 | 1 | fat | abdomen | 1 | F | 1732 | 651 | 46.76674 | 60.56582 | 73.78753 | 91.28176 | 41 | 12 | 2.3672055 | 1732 | FALSE | FALSE |
| donor1_fat.ACGGAAGCACACCTAA-1 | donor1_fat | ACGGAAGCACACCTAA-1 | 1 | fat | abdomen | 1 | F | 5069 | 2279 | 17.65634 | 25.42908 | 36.41744 | 56.77648 | 369 | 14 | 7.2795423 | 5069 | FALSE | FALSE |
| donor1_fat.ACGTAGTTCCCGAAAT-1 | donor1_fat | ACGTAGTTCCCGAAAT-1 | 1 | fat | abdomen | 1 | F | 3897 | 1246 | 36.23300 | 49.73056 | 62.63793 | 80.70310 | 39 | 11 | 1.0007698 | 3897 | FALSE | FALSE |
| donor1_fat.ACGTTCCAGATGAATC-1 | donor1_fat | ACGTTCCAGATGAATC-1 | 1 | fat | abdomen | 1 | F | 1884 | 1136 | 28.60934 | 36.57113 | 47.29299 | 66.24204 | 280 | 13 | 14.8619958 | 1884 | FALSE | FALSE |
| donor1_fat.ACTCTCGAGCCGTTGC-1 | donor1_fat | ACTCTCGAGCCGTTGC-1 | 1 | fat | abdomen | 1 | F | 1220 | 611 | 41.96721 | 53.77049 | 66.31148 | 90.90164 | 154 | 15 | 12.6229508 | 1220 | FALSE | FALSE |
| donor1_fat.ACTTCCGTCAGGAACG-1 | donor1_fat | ACTTCCGTCAGGAACG-1 | 1 | fat | abdomen | 1 | F | 4898 | 2364 | 17.29277 | 24.37730 | 34.29971 | 53.14414 | 367 | 16 | 7.4928542 | 4898 | FALSE | FALSE |
| donor1_fat.AGACACTCAGGCGATA-1 | donor1_fat | AGACACTCAGGCGATA-1 | 1 | fat | abdomen | 1 | F | 4601 | 1518 | 33.51445 | 46.70724 | 59.70441 | 76.26603 | 171 | 12 | 3.7165834 | 4601 | FALSE | FALSE |
| donor1_fat.AGACCATTCAAAGGTA-1 | donor1_fat | AGACCATTCAAAGGTA-1 | 1 | fat | abdomen | 1 | F | 7030 | 2375 | 27.28307 | 38.50640 | 50.54054 | 66.28734 | 27 | 10 | 0.3840683 | 7030 | FALSE | FALSE |
| donor1_fat.AGAGAGCCAAGCCCAC-1 | donor1_fat | AGAGAGCCAAGCCCAC-1 | 1 | fat | abdomen | 1 | F | 3437 | 1047 | 43.26447 | 57.81205 | 69.53739 | 84.08496 | 85 | 12 | 2.4730870 | 3437 | FALSE | FALSE |
| donor1_fat.AGAGCCCAGTCTAGCT-1 | donor1_fat | AGAGCCCAGTCTAGCT-1 | 1 | fat | abdomen | 1 | F | 4781 | 1680 | 33.44489 | 46.37105 | 58.12592 | 73.41560 | 110 | 14 | 2.3007739 | 4781 | FALSE | FALSE |
| donor1_fat.AGATCCAGTGCAGTGA-1 | donor1_fat | AGATCCAGTGCAGTGA-1 | 1 | fat | abdomen | 1 | F | 3119 | 1398 | 30.49054 | 42.70600 | 53.83136 | 71.20872 | 84 | 12 | 2.6931709 | 3119 | FALSE | FALSE |
| donor1_fat.AGATCGTGTTGCGTAT-1 | donor1_fat | AGATCGTGTTGCGTAT-1 | 1 | fat | abdomen | 1 | F | 4444 | 1616 | 32.06571 | 45.16202 | 56.79568 | 73.33483 | 7 | 5 | 0.1575158 | 4444 | FALSE | FALSE |
| donor1_fat.AGGAGGTCAATACGAA-1 | donor1_fat | AGGAGGTCAATACGAA-1 | 1 | fat | abdomen | 1 | F | 3179 | 1211 | 36.83548 | 49.82699 | 61.81189 | 77.63448 | 108 | 12 | 3.3972947 | 3179 | FALSE | FALSE |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| donor5_recovery.TTTCGATAGTGGAATT-1 | donor5_recovery | TTTCGATAGTGGAATT-1 | 5 | mixed | abdomen | 4 | ? | 3048 | 1575 | 21.42388 | 31.33202 | 42.84777 | 64.20604 | 139 | 16 | 4.5603675 | 3048 | FALSE | FALSE |
| donor5_recovery.TTTCGATGTGCTGATT-1 | donor5_recovery | TTTCGATGTGCTGATT-1 | 5 | mixed | abdomen | 4 | ? | 2238 | 1400 | 17.20286 | 25.24576 | 36.90795 | 59.78552 | 57 | 15 | 2.5469169 | 2238 | FALSE | FALSE |
| donor5_recovery.TTTGACTAGATGGGCT-1 | donor5_recovery | TTTGACTAGATGGGCT-1 | 5 | mixed | abdomen | 4 | ? | 2448 | 1208 | 27.69608 | 39.95098 | 53.10458 | 71.07843 | 95 | 16 | 3.8807190 | 2448 | FALSE | FALSE |
| donor5_recovery.TTTGACTAGTGTTCAC-1 | donor5_recovery | TTTGACTAGTGTTCAC-1 | 5 | mixed | abdomen | 4 | ? | 6499 | 2564 | 24.23450 | 34.06678 | 44.72996 | 60.70165 | 78 | 13 | 1.2001846 | 6499 | FALSE | FALSE |
| donor5_recovery.TTTGACTGTCCAAATC-1 | donor5_recovery | TTTGACTGTCCAAATC-1 | 5 | mixed | abdomen | 4 | ? | 958 | 673 | 24.32150 | 35.80376 | 50.62630 | 81.94154 | 8 | 4 | 0.8350731 | 958 | FALSE | FALSE |
| donor5_recovery.TTTGACTGTGATTCTG-1 | donor5_recovery | TTTGACTGTGATTCTG-1 | 5 | mixed | abdomen | 4 | ? | 1568 | 955 | 26.97704 | 37.75510 | 50.70153 | 70.98214 | 71 | 14 | 4.5280612 | 1568 | FALSE | FALSE |
| donor5_recovery.TTTGACTGTTGTCTAG-1 | donor5_recovery | TTTGACTGTTGTCTAG-1 | 5 | mixed | abdomen | 4 | ? | 936 | 593 | 29.70085 | 42.30769 | 58.01282 | 90.06410 | 38 | 11 | 4.0598291 | 936 | FALSE | FALSE |
| donor5_recovery.TTTGATCAGATGACCG-1 | donor5_recovery | TTTGATCAGATGACCG-1 | 5 | mixed | abdomen | 4 | ? | 956 | 596 | 30.75314 | 43.61925 | 58.57741 | 89.95816 | 25 | 8 | 2.6150628 | 956 | FALSE | FALSE |
| donor5_recovery.TTTGATCAGCAATAAC-1 | donor5_recovery | TTTGATCAGCAATAAC-1 | 5 | mixed | abdomen | 4 | ? | 2148 | 939 | 34.35754 | 48.51024 | 61.17318 | 79.56238 | 71 | 14 | 3.3054004 | 2148 | FALSE | FALSE |
| donor5_recovery.TTTGATCAGTTACTCG-1 | donor5_recovery | TTTGATCAGTTACTCG-1 | 5 | mixed | abdomen | 4 | ? | 3145 | 1486 | 21.90779 | 32.84579 | 46.45469 | 67.59936 | 66 | 15 | 2.0985692 | 3145 | FALSE | FALSE |
| donor5_recovery.TTTGATCGTTGGGCCT-1 | donor5_recovery | TTTGATCGTTGGGCCT-1 | 5 | mixed | abdomen | 4 | ? | 2897 | 1316 | 30.54884 | 42.18157 | 54.57370 | 71.83293 | 182 | 17 | 6.2823611 | 2897 | FALSE | FALSE |
| donor5_recovery.TTTGATCTCAAAGGTA-1 | donor5_recovery | TTTGATCTCAAAGGTA-1 | 5 | mixed | abdomen | 4 | ? | 2155 | 1136 | 26.07889 | 36.98376 | 50.06961 | 70.48724 | 98 | 14 | 4.5475638 | 2155 | FALSE | FALSE |
| donor5_recovery.TTTGGAGAGTGCAAAT-1 | donor5_recovery | TTTGGAGAGTGCAAAT-1 | 5 | mixed | abdomen | 4 | ? | 1054 | 601 | 34.91461 | 48.10247 | 61.95446 | 90.41746 | 43 | 14 | 4.0796964 | 1054 | FALSE | FALSE |
| donor5_recovery.TTTGGAGCATAGGTAA-1 | donor5_recovery | TTTGGAGCATAGGTAA-1 | 5 | mixed | abdomen | 4 | ? | 1846 | 1050 | 23.34778 | 33.91116 | 47.50813 | 70.20585 | 89 | 14 | 4.8212351 | 1846 | FALSE | FALSE |
| donor5_recovery.TTTGGAGGTCAGTTTG-1 | donor5_recovery | TTTGGAGGTCAGTTTG-1 | 5 | mixed | abdomen | 4 | ? | 1222 | 769 | 26.67758 | 38.21604 | 53.43699 | 77.98691 | 51 | 13 | 4.1734861 | 1222 | FALSE | FALSE |
| donor5_recovery.TTTGGAGGTCCAAGAG-1 | donor5_recovery | TTTGGAGGTCCAAGAG-1 | 5 | mixed | abdomen | 4 | ? | 1829 | 1058 | 27.28267 | 37.88956 | 49.64461 | 69.49153 | 28 | 12 | 1.5308912 | 1829 | FALSE | FALSE |
| donor5_recovery.TTTGGAGGTCGACTTA-1 | donor5_recovery | TTTGGAGGTCGACTTA-1 | 5 | mixed | abdomen | 4 | ? | 2389 | 1430 | 17.91545 | 26.28715 | 38.21683 | 61.07158 | 154 | 21 | 6.4462118 | 2389 | FALSE | FALSE |
| donor5_recovery.TTTGGAGGTTGAGAGC-1 | donor5_recovery | TTTGGAGGTTGAGAGC-1 | 5 | mixed | abdomen | 4 | ? | 1618 | 993 | 21.69345 | 32.26205 | 46.66255 | 69.53028 | 38 | 11 | 2.3485785 | 1618 | FALSE | FALSE |
| donor5_recovery.TTTGGAGTCCAAGAGG-1 | donor5_recovery | TTTGGAGTCCAAGAGG-1 | 5 | mixed | abdomen | 4 | ? | 1262 | 830 | 23.85103 | 34.62758 | 50.07924 | 73.85103 | 69 | 14 | 5.4675119 | 1262 | FALSE | FALSE |
| donor5_recovery.TTTGGAGTCTCAACCC-1 | donor5_recovery | TTTGGAGTCTCAACCC-1 | 5 | mixed | abdomen | 4 | ? | 1135 | 717 | 29.51542 | 40.79295 | 54.44934 | 80.88106 | 18 | 10 | 1.5859031 | 1135 | FALSE | FALSE |
| donor5_recovery.TTTGGTTAGCCTCTTC-1 | donor5_recovery | TTTGGTTAGCCTCTTC-1 | 5 | mixed | abdomen | 4 | ? | 763 | 584 | 24.90170 | 36.56619 | 49.67235 | 88.99083 | 24 | 8 | 3.1454784 | 763 | FALSE | FALSE |
| donor5_recovery.TTTGGTTCACTCTAGA-1 | donor5_recovery | TTTGGTTCACTCTAGA-1 | 5 | mixed | abdomen | 4 | ? | 812 | 570 | 27.58621 | 40.14778 | 54.43350 | 91.37931 | 24 | 8 | 2.9556650 | 812 | FALSE | FALSE |
| donor5_recovery.TTTGGTTGTACGTGAG-1 | donor5_recovery | TTTGGTTGTACGTGAG-1 | 5 | mixed | abdomen | 4 | ? | 755 | 494 | 30.99338 | 44.90066 | 61.05960 | 100.00000 | 34 | 9 | 4.5033113 | 755 | FALSE | FALSE |
| donor5_recovery.TTTGGTTGTCCTACAA-1 | donor5_recovery | TTTGGTTGTCCTACAA-1 | 5 | mixed | abdomen | 4 | ? | 835 | 625 | 24.07186 | 36.04790 | 49.10180 | 85.02994 | 27 | 6 | 3.2335329 | 835 | FALSE | FALSE |
| donor5_recovery.TTTGGTTGTGACTCTA-1 | donor5_recovery | TTTGGTTGTGACTCTA-1 | 5 | mixed | abdomen | 4 | ? | 938 | 572 | 29.63753 | 43.39019 | 60.34115 | 92.32409 | 2 | 2 | 0.2132196 | 938 | FALSE | FALSE |
| donor5_recovery.TTTGGTTTCGACCAAT-1 | donor5_recovery | TTTGGTTTCGACCAAT-1 | 5 | mixed | abdomen | 4 | ? | 1328 | 670 | 34.33735 | 48.34337 | 64.30723 | 87.19880 | 67 | 10 | 5.0451807 | 1328 | FALSE | FALSE |
| donor5_recovery.TTTGTTGGTCCTACGG-1 | donor5_recovery | TTTGTTGGTCCTACGG-1 | 5 | mixed | abdomen | 4 | ? | 795 | 552 | 28.30189 | 41.63522 | 55.72327 | 93.45912 | 44 | 12 | 5.5345912 | 795 | FALSE | FALSE |
| donor5_recovery.TTTGTTGGTCGCAGTC-1 | donor5_recovery | TTTGTTGGTCGCAGTC-1 | 5 | mixed | abdomen | 4 | ? | 1872 | 1250 | 16.18590 | 24.73291 | 37.01923 | 59.93590 | 66 | 14 | 3.5256410 | 1872 | FALSE | FALSE |
| donor5_recovery.TTTGTTGGTGGCGTAA-1 | donor5_recovery | TTTGTTGGTGGCGTAA-1 | 5 | mixed | abdomen | 4 | ? | 1928 | 931 | 33.14315 | 46.21369 | 58.97303 | 77.64523 | 51 | 13 | 2.6452282 | 1928 | FALSE | FALSE |
| donor5_recovery.TTTGTTGTCGGCATTA-1 | donor5_recovery | TTTGTTGTCGGCATTA-1 | 5 | mixed | abdomen | 4 | ? | 2091 | 1206 | 21.95122 | 32.56815 | 45.28934 | 66.23625 | 53 | 14 | 2.5346724 | 2091 | FALSE | FALSE |
with(cd_df, table(outlier_sum, outlier_detected)) outlier_detected
outlier_sum FALSE TRUE
FALSE 22274 12
TRUE 12 39options(repr.plot.width=20, repr.plot.height=20)
ggplot(cd_df, aes(sum, detected, color=outlier_sum, shape=outlier_detected)) +
geom_point(size=2) +
scale_x_log10() +
scale_y_log10() +
facet_wrap(~Sample)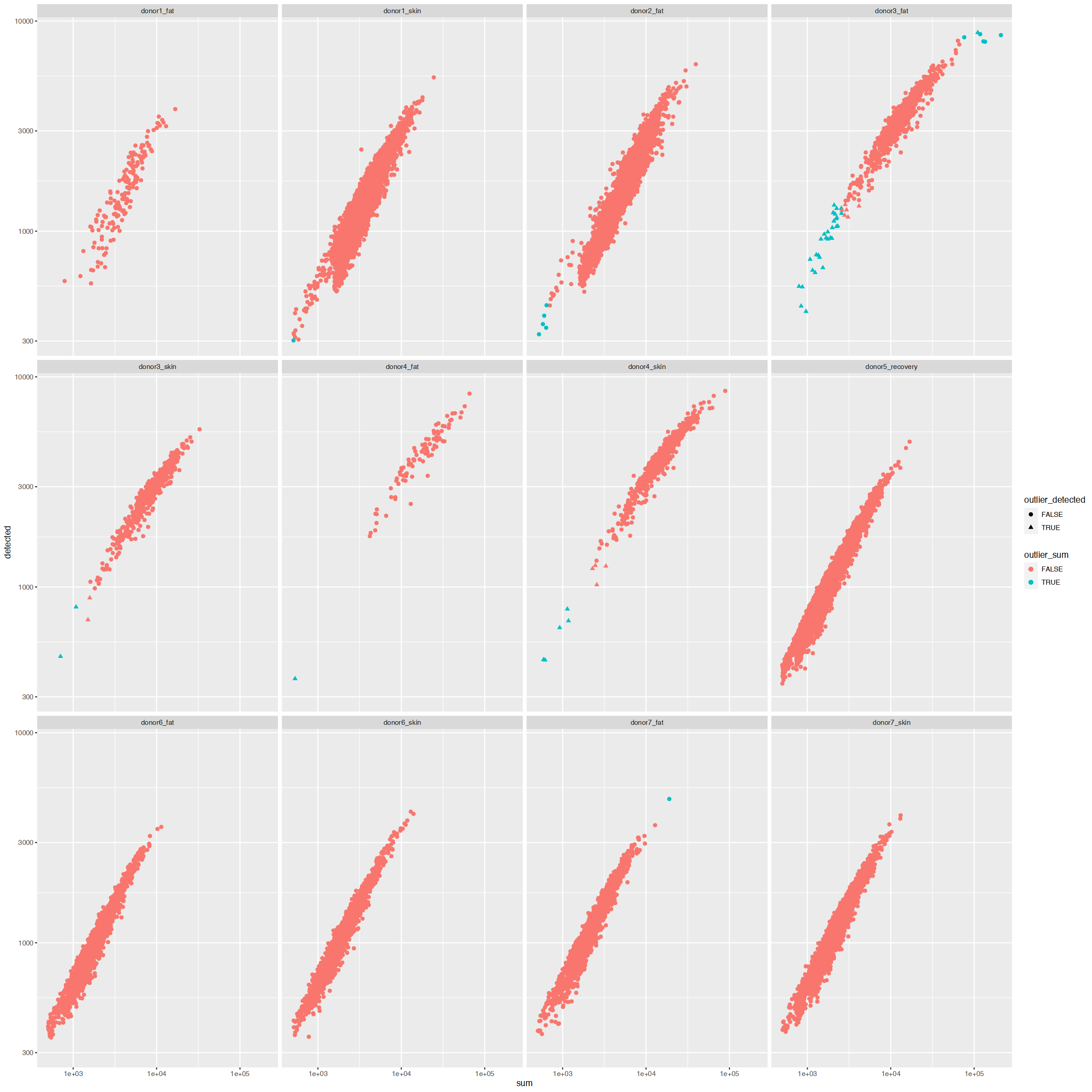
options(repr.plot.width=20, repr.plot.height=20)
ggplot(cd_df, aes(sum, subsets_Mito_percent, color=outlier_sum, shape=outlier_detected)) +
geom_point(size=2) +
scale_x_log10() +
facet_wrap(~Sample)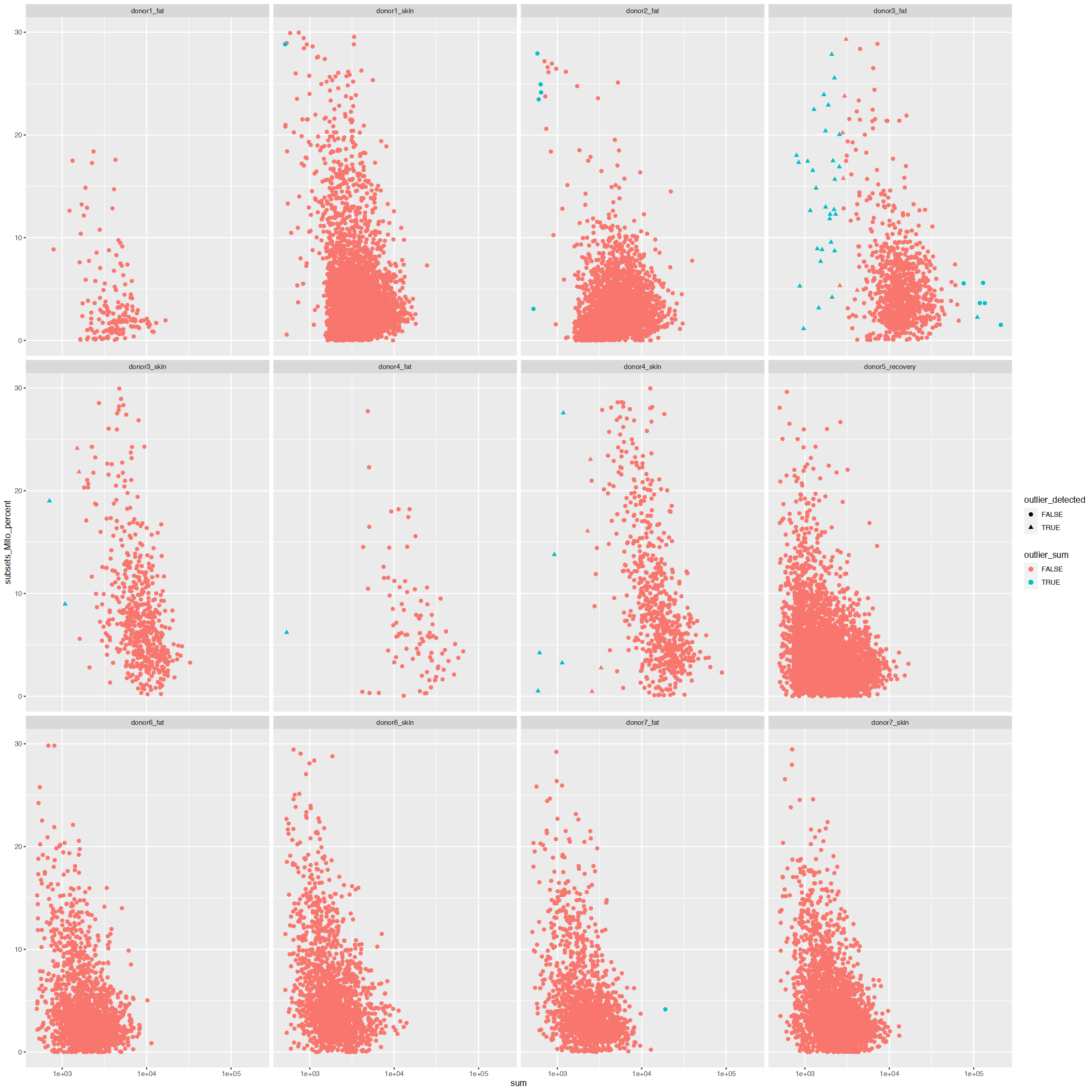
colData(sce) <- cd_df %>% DataFrame
colData(sce) DataFrame with 22337 rows and 19 columns
Sample Barcode donor
<character> <character> <numeric>
donor1_fat.AAAGAACCACATACGT-1 donor1_fat AAAGAACCACATACGT-1 1
donor1_fat.AAAGGTAGTCCGGTCA-1 donor1_fat AAAGGTAGTCCGGTCA-1 1
donor1_fat.AAAGGTATCTCTCTTC-1 donor1_fat AAAGGTATCTCTCTTC-1 1
donor1_fat.AACCACACAGTGGCTC-1 donor1_fat AACCACACAGTGGCTC-1 1
donor1_fat.AACGGGATCTGCTGAA-1 donor1_fat AACGGGATCTGCTGAA-1 1
... ... ... ...
donor5_recovery.TTTGGTTTCGACCAAT-1 donor5_recovery TTTGGTTTCGACCAAT-1 5
donor5_recovery.TTTGTTGGTCCTACGG-1 donor5_recovery TTTGTTGGTCCTACGG-1 5
donor5_recovery.TTTGTTGGTCGCAGTC-1 donor5_recovery TTTGTTGGTCGCAGTC-1 5
donor5_recovery.TTTGTTGGTGGCGTAA-1 donor5_recovery TTTGTTGGTGGCGTAA-1 5
donor5_recovery.TTTGTTGTCGGCATTA-1 donor5_recovery TTTGTTGTCGGCATTA-1 5
tissue tissue_origin sequencing_run
<character> <character> <numeric>
donor1_fat.AAAGAACCACATACGT-1 fat abdomen 1
donor1_fat.AAAGGTAGTCCGGTCA-1 fat abdomen 1
donor1_fat.AAAGGTATCTCTCTTC-1 fat abdomen 1
donor1_fat.AACCACACAGTGGCTC-1 fat abdomen 1
donor1_fat.AACGGGATCTGCTGAA-1 fat abdomen 1
... ... ... ...
donor5_recovery.TTTGGTTTCGACCAAT-1 mixed abdomen 4
donor5_recovery.TTTGTTGGTCCTACGG-1 mixed abdomen 4
donor5_recovery.TTTGTTGGTCGCAGTC-1 mixed abdomen 4
donor5_recovery.TTTGTTGGTGGCGTAA-1 mixed abdomen 4
donor5_recovery.TTTGTTGTCGGCATTA-1 mixed abdomen 4
sex sum detected
<character> <numeric> <integer>
donor1_fat.AAAGAACCACATACGT-1 F 4276 1721
donor1_fat.AAAGGTAGTCCGGTCA-1 F 3023 1084
donor1_fat.AAAGGTATCTCTCTTC-1 F 3892 1422
donor1_fat.AACCACACAGTGGCTC-1 F 7722 2633
donor1_fat.AACGGGATCTGCTGAA-1 F 3256 1284
... ... ... ...
donor5_recovery.TTTGGTTTCGACCAAT-1 ? 1328 670
donor5_recovery.TTTGTTGGTCCTACGG-1 ? 795 552
donor5_recovery.TTTGTTGGTCGCAGTC-1 ? 1872 1250
donor5_recovery.TTTGTTGGTGGCGTAA-1 ? 1928 931
donor5_recovery.TTTGTTGTCGGCATTA-1 ? 2091 1206
percent.top_50 percent.top_100
<numeric> <numeric>
donor1_fat.AAAGAACCACATACGT-1 33.8868 42.7736
donor1_fat.AAAGGTAGTCCGGTCA-1 39.6626 52.9937
donor1_fat.AAAGGTATCTCTCTTC-1 34.1213 46.5570
donor1_fat.AACCACACAGTGGCTC-1 26.8972 38.1248
donor1_fat.AACGGGATCTGCTGAA-1 34.3366 46.8059
... ... ...
donor5_recovery.TTTGGTTTCGACCAAT-1 34.3373 48.3434
donor5_recovery.TTTGTTGGTCCTACGG-1 28.3019 41.6352
donor5_recovery.TTTGTTGGTCGCAGTC-1 16.1859 24.7329
donor5_recovery.TTTGTTGGTGGCGTAA-1 33.1432 46.2137
donor5_recovery.TTTGTTGTCGGCATTA-1 21.9512 32.5681
percent.top_200 percent.top_500
<numeric> <numeric>
donor1_fat.AAAGAACCACATACGT-1 52.9935 69.3171
donor1_fat.AAAGGTAGTCCGGTCA-1 65.0017 80.6814
donor1_fat.AAAGGTATCTCTCTTC-1 58.3505 75.9250
donor1_fat.AACCACACAGTGGCTC-1 48.6791 64.1285
donor1_fat.AACGGGATCTGCTGAA-1 59.0602 75.9214
... ... ...
donor5_recovery.TTTGGTTTCGACCAAT-1 64.3072 87.1988
donor5_recovery.TTTGTTGGTCCTACGG-1 55.7233 93.4591
donor5_recovery.TTTGTTGGTCGCAGTC-1 37.0192 59.9359
donor5_recovery.TTTGTTGGTGGCGTAA-1 58.9730 77.6452
donor5_recovery.TTTGTTGTCGGCATTA-1 45.2893 66.2363
subsets_Mito_sum subsets_Mito_detected
<numeric> <integer>
donor1_fat.AAAGAACCACATACGT-1 752 16
donor1_fat.AAAGGTAGTCCGGTCA-1 52 12
donor1_fat.AAAGGTATCTCTCTTC-1 48 12
donor1_fat.AACCACACAGTGGCTC-1 103 13
donor1_fat.AACGGGATCTGCTGAA-1 123 11
... ... ...
donor5_recovery.TTTGGTTTCGACCAAT-1 67 10
donor5_recovery.TTTGTTGGTCCTACGG-1 44 12
donor5_recovery.TTTGTTGGTCGCAGTC-1 66 14
donor5_recovery.TTTGTTGGTGGCGTAA-1 51 13
donor5_recovery.TTTGTTGTCGGCATTA-1 53 14
subsets_Mito_percent total
<numeric> <numeric>
donor1_fat.AAAGAACCACATACGT-1 17.58653 4276
donor1_fat.AAAGGTAGTCCGGTCA-1 1.72015 3023
donor1_fat.AAAGGTATCTCTCTTC-1 1.23330 3892
donor1_fat.AACCACACAGTGGCTC-1 1.33385 7722
donor1_fat.AACGGGATCTGCTGAA-1 3.77764 3256
... ... ...
donor5_recovery.TTTGGTTTCGACCAAT-1 5.04518 1328
donor5_recovery.TTTGTTGGTCCTACGG-1 5.53459 795
donor5_recovery.TTTGTTGGTCGCAGTC-1 3.52564 1872
donor5_recovery.TTTGTTGGTGGCGTAA-1 2.64523 1928
donor5_recovery.TTTGTTGTCGGCATTA-1 2.53467 2091
outlier_sum outlier_detected
<outlier.filter> <outlier.filter>
donor1_fat.AAAGAACCACATACGT-1 FALSE FALSE
donor1_fat.AAAGGTAGTCCGGTCA-1 FALSE FALSE
donor1_fat.AAAGGTATCTCTCTTC-1 FALSE FALSE
donor1_fat.AACCACACAGTGGCTC-1 FALSE FALSE
donor1_fat.AACGGGATCTGCTGAA-1 FALSE FALSE
... ... ...
donor5_recovery.TTTGGTTTCGACCAAT-1 FALSE FALSE
donor5_recovery.TTTGTTGGTCCTACGG-1 FALSE FALSE
donor5_recovery.TTTGTTGGTCGCAGTC-1 FALSE FALSE
donor5_recovery.TTTGTTGGTGGCGTAA-1 FALSE FALSE
donor5_recovery.TTTGTTGTCGGCATTA-1 FALSE FALSEsce <- sce[,!colData(sce)$outlier_detected]
sce <- sce[,!colData(sce)$outlier_sum]
sceclass: SingleCellExperiment
dim: 18627 22274
metadata(0):
assays(1): counts
rownames(18627): ENSG00000187634.SAMD11 ENSG00000188976.NOC2L ...
ENSG00000278817.ENSG00000278817 ENSG00000277196.ENSG00000277196
rowData names(3): ID Symbol Type
colnames(22274): donor1_fat.AAAGAACCACATACGT-1
donor1_fat.AAAGGTAGTCCGGTCA-1 ... donor5_recovery.TTTGTTGGTGGCGTAA-1
donor5_recovery.TTTGTTGTCGGCATTA-1
colData names(19): Sample Barcode ... outlier_sum outlier_detected
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):saveRDS(sce, file="../output/0.1-sce-after-filtering.rds")markers <- googlesheets4::read_sheet(md_sheet, sheet = "markers")
markers✔ Reading from halin-bauer-hLECs-metadata.
✔ Range ''markers''.
| gene_name | subpopulation | subpopshort | direction | known |
|---|---|---|---|---|
| <chr> | <chr> | <chr> | <chr> | <chr> |
| ENSG00000117707.PROX1 | LECs | LEC | positive | yes |
| ENSG00000162493.PDPN | LECs | LEC | positive | yes |
| ENSG00000133800.LYVE1 | capillary-LEC | cpLEC | positive | yes |
| ENSG00000137077.CCL21 | capillary-LEC | cpLEC | positive | yes |
| ENSG00000189056.RELN | capillary-LEC | cpLEC | positive | yes |
| ENSG00000129048.ACKR4 | capillary-LEC | cpLEC | negative | yes |
| ENSG00000108691.CCL2 | capillary-LEC | cpLEC | negative | no |
| ENSG00000081041.CXCL2 | capillary-LEC | cpLEC | negative | no |
| ENSG00000133800.LYVE1 | precollector-LEC | pcLEC | positive | yes |
| ENSG00000137077.CCL21 | precollector-LEC | pcLEC | positive | yes |
| ENSG00000189056.RELN | precollector-LEC | pcLEC | positive | yes |
| ENSG00000129048.ACKR4 | precollector-LEC | pcLEC | negative | yes |
| ENSG00000108691.CCL2 | precollector-LEC | pcLEC | positive | no |
| ENSG00000081041.CXCL2 | precollector-LEC | pcLEC | positive | no |
| ENSG00000133800.LYVE1 | collector-LEC | clLEC | negative | yes |
| ENSG00000137077.CCL21 | collector-LEC | clLEC | negative | yes |
| ENSG00000189056.RELN | collector-LEC | clLEC | negative | yes |
| ENSG00000129048.ACKR4 | collector-LEC | clLEC | positive | yes |
| ENSG00000108691.CCL2 | collector-LEC | clLEC | positive | no |
| ENSG00000081041.CXCL2 | collector-LEC | clLEC | positive | no |
| ENSG00000137077.CCL21 | valve-LEC | vLEC | negative | yes |
| ENSG00000133800.LYVE1 | valve-LEC | vLEC | negative | yes |
| ENSG00000013297.CLDN11 | valve-LEC | vLEC | positive | yes |
| ENSG00000272398.CD24 | valve-LEC | vLEC | positive | no |
| ENSG00000149564.ESAM | valve-LEC | vLEC | positive | no |
| ENSG00000170017.ALCAM | valve-LEC | vLEC | positive | no |
| ENSG00000108821.COL1A1 | contaminant-fibroblasts-smooth-muscle | CONT-FSM | positive | yes |
| ENSG00000113721.PDGFRB | contaminant-fibroblasts-smooth-muscle | CONT-FSM | positive | yes |
| ENSG00000107796.ACTA2 | contaminant-fibroblasts-smooth-muscle | CONT-FSM | positive | yes |
| ENSG00000186081.KRT5 | contaminant-keratinocytes | CONT-K | positive | yes |
| ENSG00000186847.KRT14 | contaminant-keratinocytes | CONT-K | positive | yes |
| ENSG00000081237.PTPRC | contaminant-leukocytes | CONT-L | positive | yes |
| ENSG00000102755.FLT1 | contaminant-blood-endothelial-cells | CONT-BEC | positive | yes |
| ENSG00000154721.JAM2 | contaminant-blood-endothelial-cells | CONT-BEC | positive | yes |
| ENSG00000090339.ICAM1 | LECs | LEC | positive | ? |
| ENSG00000103335.PIEZO1 | LECs | LEC | positive | ? |
| ENSG00000154864.PIEZO2 | LECs | LEC | positive | ? |
sce <- logNormCounts(sce)
sceclass: SingleCellExperiment
dim: 18627 22274
metadata(0):
assays(2): counts logcounts
rownames(18627): ENSG00000187634.SAMD11 ENSG00000188976.NOC2L ...
ENSG00000278817.ENSG00000278817 ENSG00000277196.ENSG00000277196
rowData names(3): ID Symbol Type
colnames(22274): donor1_fat.AAAGAACCACATACGT-1
donor1_fat.AAAGGTAGTCCGGTCA-1 ... donor5_recovery.TTTGTTGGTGGCGTAA-1
donor5_recovery.TTTGTTGTCGGCATTA-1
colData names(20): Sample Barcode ... outlier_detected sizeFactor
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):mgv <- modelGeneVar(sce, block=sce$Sample)
per_block <- mgv$per.block
mgv$mgv$per.block <- NULL
per_blockDataFrame with 18627 rows and 12 columns
donor1_fat
<DataFrame>
1 0.0000000:0.0000000:0.0000000:...
2 0.0447544:0.0483622:0.0471062:...
3 0.0299111:0.0420068:0.0315344:...
4 0.0000000:0.0000000:0.0000000:...
5 0.0000000:0.0000000:0.0000000:...
... ...
18623 0.00527968:0.00440426:0.00557591:...
18624 0.00000000:0.00000000:0.00000000:...
18625 0.00000000:0.00000000:0.00000000:...
18626 0.01379355:0.01574627:0.01456116:...
18627 0.00000000:0.00000000:0.00000000:...
donor1_skin
<DataFrame>
1 0.000192091:0.000140327:0.000210392:...
2 0.030096853:0.034786115:0.032963547:...
3 0.007230149:0.009100637:0.007918963:...
4 0.000673060:0.000577614:0.000737182:...
5 0.000000000:0.000000000:0.000000000:...
... ...
18623 0.005793431:0.006536338:0.006345371:...
18624 0.000655845:0.000823243:0.000718328:...
18625 0.000000000:0.000000000:0.000000000:...
18626 0.015571469:0.017150311:0.017054905:...
18627 0.000000000:0.000000000:0.000000000:...
donor2_fat
<DataFrame>
1 0.000257818:9.52261e-05:0.000231155:...
2 0.030989651:2.27018e-02:0.027517226:...
3 0.000541784:4.51619e-04:0.000485743:...
4 0.001016535:7.51721e-04:0.000911335:...
5 0.000000000:0.00000e+00:0.000000000:...
... ...
18623 0.006846574:0.00432770:0.006130807:...
18624 0.063328584:0.05779895:0.055368734:...
18625 0.000000000:0.00000000:0.000000000:...
18626 0.018603745:0.01511799:0.016598744:...
18627 0.000612517:0.00105725:0.000549155:...
donor3_fat
<DataFrame>
1 0.005251594:0.002826777:0.002720376:...
2 0.058227092:0.025883220:0.030161476:...
3 0.002911593:0.001252882:0.001508233:...
4 0.000463204:0.000109945:0.000239944:...
5 0.000768523:0.000603030:0.000398102:...
... ...
18623 0.01701294:0.007165761:0.008812857:...
18624 0.04656016:0.019818419:0.024118293:...
18625 0.00047452:0.000119722:0.000245806:...
18626 0.02401902:0.009052763:0.012442053:...
18627 0.00000000:0.000000000:0.000000000:...
donor3_skin
<DataFrame>
1 0.000000000:0.000000000:0.000000000:...
2 0.077795605:0.048279909:0.058675308:...
3 0.004722309:0.005066520:0.003738193:...
4 0.000962391:0.000527005:0.000763779:...
5 0.000000000:0.000000000:0.000000000:...
... ...
18623 0.03084809:0.02389855:0.02399428:...
18624 0.05316327:0.03602979:0.04074557:...
18625 0.00000000:0.00000000:0.00000000:...
18626 0.00585365:0.00409068:0.00463021:...
18627 0.00000000:0.00000000:0.00000000:...
donor4_fat
<DataFrame>
1 0.01069955:3.53034e-03:0.004835728:...
2 0.09387548:4.16551e-02:0.042425119:...
3 0.01530288:6.05392e-03:0.006916231:...
4 0.00104185:9.55201e-05:0.000470872:...
5 0.00000000:0.00000e+00:0.000000000:...
... ...
18623 0.0202734:0.00534214:0.00916268:...
18624 0.0624849:0.02416214:0.02823992:...
18625 0.0000000:0.00000000:0.00000000:...
18626 0.0377530:0.01659354:0.01706262:...
18627 0.0000000:0.00000000:0.00000000:...
donor4_skin
<DataFrame>
1 0.001684255:0.000984743:0.000795111:...
2 0.099895592:0.041682785:0.047077413:...
3 0.018399329:0.011478903:0.008685330:...
4 0.002311079:0.000617455:0.001091023:...
5 0.000486684:0.000148986:0.000229756:...
... ...
18623 0.03531342:0.017654658:0.016666491:...
18624 0.08061596:0.029509405:0.038012706:...
18625 0.00000000:0.000000000:0.000000000:...
18626 0.00619706:0.002728810:0.002925505:...
18627 0.00156129:0.000797139:0.000737061:...
donor5_recovery
<DataFrame>
1 0.002386149:0.003595803:0.004200088:...
2 0.036177813:0.066393947:0.063676351:...
3 0.005542531:0.010860622:0.009755932:...
4 0.000470654:0.000546747:0.000828443:...
5 0.000000000:0.000000000:0.000000000:...
... ...
18623 0.00385830:0.00629160:0.00679135:...
18624 0.00000000:0.00000000:0.00000000:...
18625 0.00000000:0.00000000:0.00000000:...
18626 0.00218717:0.00460785:0.00384985:...
18627 0.00000000:0.00000000:0.00000000:...
donor6_fat donor6_skin
<DataFrame> <DataFrame>
1 0.00000000:0.00000000:0.00000000:... 0.00116394:0.00130381:0.00192853:...
2 0.02855809:0.04743540:0.04610573:... 0.03484125:0.06236383:0.05771629:...
3 0.00443248:0.00746152:0.00715685:... 0.00171647:0.00298939:0.00284401:...
4 0.00105621:0.00222445:0.00170539:... 0.00000000:0.00000000:0.00000000:...
5 0.00000000:0.00000000:0.00000000:... 0.00000000:0.00000000:0.00000000:...
... ... ...
18623 0.00672034:0.0142582:0.01085087:... 0.01009389:0.0186100:0.01672428:...
18624 0.00000000:0.0000000:0.00000000:... 0.00000000:0.0000000:0.00000000:...
18625 0.00000000:0.0000000:0.00000000:... 0.00000000:0.0000000:0.00000000:...
18626 0.00103449:0.0021339:0.00167032:... 0.00490375:0.0084261:0.00812499:...
18627 0.00000000:0.0000000:0.00000000:... 0.00000000:0.0000000:0.00000000:...
donor7_fat
<DataFrame>
1 0.00162012:0.00274062:0.00245109:...
2 0.03856859:0.06085669:0.05814055:...
3 0.01021691:0.01666630:0.01544960:...
4 0.00108433:0.00200704:0.00164052:...
5 0.00000000:0.00000000:0.00000000:...
... ...
18623 0.012511800:0.017865490:0.01891639:...
18624 0.000000000:0.000000000:0.00000000:...
18625 0.000000000:0.000000000:0.00000000:...
18626 0.000761895:0.000990887:0.00115271:...
18627 0.000000000:0.000000000:0.00000000:...
donor7_skin
<DataFrame>
1 0.000377092:0.000412090:0.000554036:...
2 0.029380808:0.047142481:0.043156883:...
3 0.004676678:0.007048741:0.006871096:...
4 0.002052253:0.002850248:0.003015239:...
5 0.000457449:0.000606435:0.000672101:...
... ...
18623 0.00951234:0.01478812:0.01397550:...
18624 0.00000000:0.00000000:0.00000000:...
18625 0.00000000:0.00000000:0.00000000:...
18626 0.00246677:0.00309163:0.00362426:...
18627 0.00000000:0.00000000:0.00000000:...mgv <- mgv %>% as.data.frame %>% rownames_to_column("gene_name") %>% left_join(markers, multiple = "first")
mgvJoining with `by = join_by(gene_name)`| gene_name | mean | total | tech | bio | p.value | FDR | per.block.donor1_fat.mean | per.block.donor1_fat.total | per.block.donor1_fat.tech | ⋯ | per.block.donor7_skin.mean | per.block.donor7_skin.total | per.block.donor7_skin.tech | per.block.donor7_skin.bio | per.block.donor7_skin.p.value | per.block.donor7_skin.FDR | subpopulation | subpopshort | direction | known |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | ⋯ | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <chr> | <chr> | <chr> | <chr> |
| ENSG00000187634.SAMD11 | 0.0019693835 | 0.0013024782 | 0.0014938755 | -1.913973e-04 | 8.737032e-01 | 1.000000e+00 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0003770917 | 0.0004120903 | 0.0005540363 | -1.419460e-04 | 9.994747e-01 | 1.000000e+00 | NA | NA | NA | NA |
| ENSG00000188976.NOC2L | 0.0502634315 | 0.0456286194 | 0.0462268405 | -5.982211e-04 | 5.299364e-01 | 9.355400e-01 | 0.044754353 | 0.048362162 | 0.047106190 | ⋯ | 0.0293808083 | 0.0471424812 | 0.0431568827 | 3.985598e-03 | 1.187842e-01 | 5.326614e-01 | NA | NA | NA | NA |
| ENSG00000187961.KLHL17 | 0.0088003534 | 0.0100364902 | 0.0085720454 | 1.464445e-03 | 1.356136e-02 | 5.041921e-02 | 0.029911128 | 0.042006834 | 0.031534370 | ⋯ | 0.0046766780 | 0.0070487413 | 0.0068710959 | 1.776454e-04 | 3.704551e-01 | 7.408704e-01 | NA | NA | NA | NA |
| ENSG00000187583.PLEKHN1 | 0.0009276301 | 0.0008589782 | 0.0009503105 | -9.133234e-05 | 7.086989e-02 | 2.073134e-01 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0020522533 | 0.0028502483 | 0.0030152388 | -1.649905e-04 | 7.579746e-01 | 8.954704e-01 | NA | NA | NA | NA |
| ENSG00000187642.PERM1 | 0.0001427214 | 0.0001132043 | 0.0001083299 | 4.874395e-06 | 1.791375e-01 | 4.312149e-01 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0004574494 | 0.0006064354 | 0.0006721007 | -6.566524e-05 | 8.942605e-01 | 9.949778e-01 | NA | NA | NA | NA |
| ENSG00000188290.HES4 | 0.4317683304 | 0.4744427877 | 0.4318170816 | 4.262571e-02 | 1.769217e-02 | 6.342516e-02 | 0.579772219 | 0.530487385 | 0.524572804 | ⋯ | 0.5032418551 | 0.7325884217 | 0.6744572788 | 5.813114e-02 | 1.351696e-01 | 5.618877e-01 | NA | NA | NA | NA |
| ENSG00000187608.ISG15 | 0.1907224467 | 0.2125479814 | 0.1605618632 | 5.198612e-02 | 8.298437e-13 | 1.131179e-11 | 0.116807731 | 0.111024323 | 0.121563029 | ⋯ | 0.1162945309 | 0.1917556701 | 0.1700318967 | 2.172377e-02 | 5.113300e-02 | 3.398987e-01 | NA | NA | NA | NA |
| ENSG00000188157.AGRN | 0.1114768158 | 0.1148564437 | 0.1092542055 | 5.602238e-03 | 1.795969e-01 | 4.320410e-01 | 0.138418928 | 0.130595390 | 0.143518295 | ⋯ | 0.1001736813 | 0.1677008352 | 0.1467709611 | 2.092987e-02 | 3.409516e-02 | 2.628374e-01 | NA | NA | NA | NA |
| ENSG00000131591.C1orf159 | 0.0313450869 | 0.0341074411 | 0.0326217764 | 1.485665e-03 | 1.950322e-01 | 4.590749e-01 | 0.014560216 | 0.016643509 | 0.015369731 | ⋯ | 0.0290359040 | 0.0439328414 | 0.0426504987 | 1.282343e-03 | 3.502968e-01 | 7.360048e-01 | NA | NA | NA | NA |
| ENSG00000162571.TTLL10 | 0.0008546147 | 0.0005900526 | 0.0005168317 | 7.322091e-05 | 1.540465e-10 | 1.746455e-09 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0008199698 | 0.0019484717 | 0.0012047281 | 7.437436e-04 | 1.447307e-15 | 1.961519e-13 | NA | NA | NA | NA |
| ENSG00000186891.TNFRSF18 | 0.0058911157 | 0.0033663407 | 0.0034570205 | -9.067979e-05 | 1.715891e-07 | 1.429371e-06 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0006670706 | 0.0006459384 | 0.0009800834 | -3.341450e-04 | 9.999935e-01 | 1.000000e+00 | NA | NA | NA | NA |
| ENSG00000186827.TNFRSF4 | 0.1043514102 | 0.1025507891 | 0.0915769314 | 1.097386e-02 | 4.067668e-02 | 1.298149e-01 | 0.118148494 | 0.138295830 | 0.122931497 | ⋯ | 0.0535097343 | 0.0819809698 | 0.0785558824 | 3.425087e-03 | 2.885551e-01 | 7.098456e-01 | NA | NA | NA | NA |
| ENSG00000078808.SDF4 | 0.2212428288 | 0.1731091994 | 0.1900223305 | -1.691313e-02 | 9.752666e-01 | 1.000000e+00 | 0.125266097 | 0.109117598 | 0.130182344 | ⋯ | 0.1523546501 | 0.2205261063 | 0.2216055398 | -1.079434e-03 | 5.248361e-01 | 7.857325e-01 | NA | NA | NA | NA |
| ENSG00000176022.B3GALT6 | 0.0331810618 | 0.0290155465 | 0.0298696351 | -8.540886e-04 | 2.808728e-01 | 6.080890e-01 | 0.033773789 | 0.043187394 | 0.035592796 | ⋯ | 0.0172146779 | 0.0291051117 | 0.0252902950 | 3.814817e-03 | 2.685835e-02 | 2.274809e-01 | NA | NA | NA | NA |
| ENSG00000184163.C1QTNF12 | 0.0021494577 | 0.0015139299 | 0.0013974834 | 1.164465e-04 | 1.620399e-01 | 3.997888e-01 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000160087.UBE2J2 | 0.1039678327 | 0.0900077789 | 0.0929954030 | -2.987624e-03 | 6.280412e-01 | 1.000000e+00 | 0.095448125 | 0.074176088 | 0.099676791 | ⋯ | 0.0682506326 | 0.1080415891 | 0.1001470086 | 7.894581e-03 | 1.566890e-01 | 5.931810e-01 | NA | NA | NA | NA |
| ENSG00000162572.SCNN1D | 0.0039239679 | 0.0045575486 | 0.0040416201 | 5.159285e-04 | 4.918952e-08 | 4.365540e-07 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0057684617 | 0.0101547219 | 0.0084751452 | 1.679577e-03 | 5.630535e-03 | 7.276488e-02 | NA | NA | NA | NA |
| ENSG00000131584.ACAP3 | 0.0229812853 | 0.0232988062 | 0.0211932916 | 2.105515e-03 | 1.937648e-05 | 1.234214e-04 | 0.011195668 | 0.019804193 | 0.011820538 | ⋯ | 0.0194500131 | 0.0379595830 | 0.0285735981 | 9.385985e-03 | 1.328671e-05 | 4.117809e-04 | NA | NA | NA | NA |
| ENSG00000169972.PUSL1 | 0.0148668264 | 0.0123331169 | 0.0130527191 | -7.196022e-04 | 8.753588e-01 | 1.000000e+00 | 0.022273057 | 0.021196167 | 0.023497957 | ⋯ | 0.0099100098 | 0.0157438677 | 0.0145597274 | 1.184140e-03 | 1.491395e-01 | 5.811430e-01 | NA | NA | NA | NA |
| ENSG00000127054.INTS11 | 0.0863434745 | 0.0693228612 | 0.0735482434 | -4.225382e-03 | 3.658966e-01 | 7.358698e-01 | 0.084655755 | 0.072026480 | 0.088577214 | ⋯ | 0.0394623773 | 0.0632514002 | 0.0579543345 | 5.297066e-03 | 1.212164e-01 | 5.363840e-01 | NA | NA | NA | NA |
| ENSG00000224051.CPTP | 0.0437399735 | 0.0345153310 | 0.0355729713 | -1.057640e-03 | 2.192897e-01 | 5.029921e-01 | 0.028106629 | 0.021103972 | 0.029637060 | ⋯ | 0.0200511600 | 0.0345908846 | 0.0294565347 | 5.134350e-03 | 1.290151e-02 | 1.357971e-01 | NA | NA | NA | NA |
| ENSG00000169962.TAS1R3 | 0.0004052785 | 0.0005136642 | 0.0004490183 | 6.464582e-05 | 6.063849e-10 | 6.566403e-09 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000107404.DVL1 | 0.0546596741 | 0.0559125190 | 0.0546148119 | 1.297707e-03 | 1.169168e-01 | 3.097954e-01 | 0.051992919 | 0.051285625 | 0.054674802 | ⋯ | 0.0494111753 | 0.0774444246 | 0.0725473686 | 4.897056e-03 | 1.939918e-01 | 6.394011e-01 | NA | NA | NA | NA |
| ENSG00000162576.MXRA8 | 0.0733304526 | 0.0816338793 | 0.0424210381 | 3.921284e-02 | 8.369574e-10 | 8.943419e-09 | 0.041612054 | 0.048939431 | 0.043815198 | ⋯ | 0.0070240558 | 0.0104407885 | 0.0103198435 | 1.209450e-04 | 4.404284e-01 | 7.639509e-01 | NA | NA | NA | NA |
| ENSG00000175756.AURKAIP1 | 0.2877332314 | 0.2267634099 | 0.2455746749 | -1.881126e-02 | 9.404859e-01 | 1.000000e+00 | 0.358315318 | 0.322001177 | 0.351057560 | ⋯ | 0.1725839830 | 0.2569559693 | 0.2502336724 | 6.722297e-03 | 3.655864e-01 | 7.396080e-01 | NA | NA | NA | NA |
| ENSG00000221978.CCNL2 | 0.1554273129 | 0.1607440768 | 0.1474084468 | 1.333563e-02 | 5.406649e-02 | 1.654175e-01 | 0.143807887 | 0.203728633 | 0.148957683 | ⋯ | 0.1123157102 | 0.1831533488 | 0.1643020608 | 1.885129e-02 | 7.113931e-02 | 4.110676e-01 | NA | NA | NA | NA |
| ENSG00000242485.MRPL20 | 0.4041161319 | 0.3611990674 | 0.3758276205 | -1.462855e-02 | 9.394156e-01 | 1.000000e+00 | 0.435379963 | 0.374452458 | 0.415710684 | ⋯ | 0.3693702802 | 0.5208705603 | 0.5140860481 | 6.784512e-03 | 4.329849e-01 | 7.621041e-01 | NA | NA | NA | NA |
| ENSG00000235098.ANKRD65 | 0.0075721639 | 0.0037111374 | 0.0045934832 | -8.823459e-04 | 7.679673e-01 | 1.000000e+00 | 0.005096231 | 0.004103508 | 0.005382204 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000205116.TMEM88B | 0.0003453538 | 0.0001663211 | 0.0002435159 | -7.719476e-05 | 9.989500e-01 | 1.000000e+00 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0002774246 | 0.0002230429 | 0.0004076020 | -1.845591e-04 | 1.000000e+00 | 1.000000e+00 | NA | NA | NA | NA |
| ENSG00000179403.VWA1 | 0.0201312569 | 0.0184182043 | 0.0144274781 | 3.990726e-03 | 9.560126e-06 | 6.380166e-05 | 0.011154903 | 0.007099697 | 0.011777524 | ⋯ | 0.0078463236 | 0.0130163670 | 0.0115278913 | 1.488476e-03 | 4.933709e-02 | 3.325438e-01 | NA | NA | NA | NA |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋱ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| ENSG00000182484.WASH6P | 1.060511e-01 | 1.051630e-01 | 1.012401e-01 | 3.922887e-03 | 5.385302e-02 | 1.649027e-01 | 0.089568204 | 0.108006196 | 0.093636322 | ⋯ | 0.0983430607 | 0.1563313631 | 0.1441048272 | 1.222654e-02 | 1.389424e-01 | 0.567558120 | NA | NA | NA | NA |
| ENSG00000129824.RPS4Y1 | 2.235694e-01 | 2.838755e-01 | 2.897254e-01 | -5.849912e-03 | 1.472026e-03 | 6.816781e-03 | 0.002959566 | 0.001383926 | 0.003125851 | ⋯ | 0.0170548138 | 0.0324826172 | 0.0250554751 | 7.427142e-03 | 7.501188e-05 | 0.001922062 | NA | NA | NA | NA |
| ENSG00000067646.ZFY | 3.808236e-02 | 4.753073e-02 | 4.488257e-02 | 2.648161e-03 | 5.600530e-03 | 2.288586e-02 | 0.062956984 | 0.069570622 | 0.066102981 | ⋯ | 0.0111761192 | 0.0168789885 | 0.0164197643 | 4.592242e-04 | 3.602910e-01 | 0.738736203 | NA | NA | NA | NA |
| ENSG00000099715.PCDH11Y | 8.276551e-03 | 1.038031e-02 | 8.746170e-03 | 1.634137e-03 | 2.348474e-03 | 1.046374e-02 | 0.018789257 | 0.035260608 | 0.019828012 | ⋯ | 0.0056077484 | 0.0089139084 | 0.0082390261 | 6.748823e-04 | 1.474144e-01 | 0.578190854 | NA | NA | NA | NA |
| ENSG00000099721.AMELY | 3.799039e-04 | 3.942317e-04 | 3.744746e-04 | 1.975718e-05 | 2.299880e-04 | 1.239469e-03 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000092377.TBL1Y | 6.918795e-04 | 7.768121e-04 | 8.532257e-04 | -7.641360e-05 | 4.001967e-01 | 7.810283e-01 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0009483741 | 0.0013166584 | 0.0013933841 | -7.672566e-05 | 7.593519e-01 | 0.896580882 | NA | NA | NA | NA |
| ENSG00000114374.USP9Y | 3.812034e-02 | 6.343246e-02 | 6.151179e-02 | 1.920676e-03 | 8.991925e-01 | 1.000000e+00 | 0.004961967 | 0.003890137 | 0.005240431 | ⋯ | 0.0014691287 | 0.0021249447 | 0.0021584939 | -3.354920e-05 | 5.787820e-01 | 0.804795343 | NA | NA | NA | NA |
| ENSG00000067048.DDX3Y | 9.987608e-02 | 1.538440e-01 | 1.521439e-01 | 1.700035e-03 | 3.401029e-04 | 1.767338e-03 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0067399916 | 0.0129023438 | 0.0099025033 | 2.999841e-03 | 5.346960e-05 | 0.001428091 | NA | NA | NA | NA |
| ENSG00000183878.UTY | 6.276576e-02 | 1.035719e-01 | 9.944009e-02 | 4.131834e-03 | 2.371132e-05 | 1.491927e-04 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0037955497 | 0.0055075356 | 0.0055765314 | -6.899581e-05 | 5.628633e-01 | 0.797561478 | NA | NA | NA | NA |
| ENSG00000154620.TMSB4Y | 3.161534e-03 | 5.812091e-03 | 5.249932e-03 | 5.621585e-04 | 3.716597e-03 | 1.584561e-02 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000165246.NLGN4Y | 2.889787e-02 | 4.648592e-02 | 4.439543e-02 | 2.090492e-03 | 2.261591e-05 | 1.426383e-04 | 0.007101396 | 0.007967912 | 0.007499287 | ⋯ | 0.0059726097 | 0.0094529158 | 0.0087750778 | 6.778379e-04 | 1.616008e-01 | 0.599288583 | NA | NA | NA | NA |
| ENSG00000172468.HSFY1 | 1.774793e-04 | 1.349566e-04 | 1.911604e-04 | -5.620376e-05 | 9.754492e-01 | 1.000000e+00 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0002446283 | 0.0001734250 | 0.0003594165 | -1.859914e-04 | 1.000000e+00 | 1.000000000 | NA | NA | NA | NA |
| ENSG00000169953.HSFY2 | 2.195080e-04 | 2.132608e-04 | 2.215071e-04 | -8.246239e-06 | 6.838196e-01 | 1.000000e+00 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0002892259 | 0.0002424224 | 0.0004249408 | -1.825184e-04 | 1.000000e+00 | 1.000000000 | NA | NA | NA | NA |
| ENSG00000012817.KDM5D | 1.168414e-02 | 2.019782e-02 | 1.928814e-02 | 9.096875e-04 | 2.116818e-03 | 9.515955e-03 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000198692.EIF1AY | 4.603123e-02 | 7.533155e-02 | 7.455884e-02 | 7.727028e-04 | 9.981512e-02 | 2.736453e-01 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0009624456 | 0.0009776989 | 0.0014140584 | -4.363595e-04 | 9.999604e-01 | 1.000000000 | NA | NA | NA | NA |
| ENSG00000280969.RPS4Y2 | 3.188270e-05 | 2.432296e-05 | 5.147908e-05 | -2.715612e-05 | 1.000000e+00 | 1.000000e+00 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000169807.PRY2 | 6.112707e-05 | 5.399550e-05 | 8.281698e-05 | -2.882148e-05 | 8.976356e-01 | 1.000000e+00 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000187191.DAZ3 | 2.244223e-05 | 1.703156e-05 | 2.012135e-05 | -3.089796e-06 | 8.264456e-01 | 1.000000e+00 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000205916.DAZ4 | 1.205705e-04 | 2.539327e-04 | 1.831278e-04 | 7.080490e-05 | 7.884693e-07 | 6.082992e-06 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000278704.ENSG00000278704 | 6.738617e-03 | 7.672991e-03 | 7.311764e-03 | 3.612269e-04 | 3.103076e-01 | 6.540698e-01 | 0.007461762 | 0.008797108 | 0.007879719 | ⋯ | 0.0070398383 | 0.0101438386 | 0.0103430307 | -1.991921e-04 | 5.972746e-01 | 0.811804880 | NA | NA | NA | NA |
| ENSG00000277400.ENSG00000277400 | 3.346039e-03 | 3.604205e-03 | 3.321011e-03 | 2.831942e-04 | 6.867569e-01 | 1.000000e+00 | 0.009749444 | 0.015018162 | 0.010294400 | ⋯ | 0.0003640715 | 0.0003841243 | 0.0005349066 | -1.507823e-04 | 9.998439e-01 | 1.000000000 | NA | NA | NA | NA |
| ENSG00000274847.MAFIP | 2.747836e-02 | 3.523302e-02 | 3.322526e-02 | 2.007761e-03 | 1.097189e-01 | 2.950113e-01 | 0.045624753 | 0.054548175 | 0.048017210 | ⋯ | 0.0266886287 | 0.0400806811 | 0.0392040406 | 8.766406e-04 | 3.874484e-01 | 0.746886890 | NA | NA | NA | NA |
| ENSG00000276256.ENSG00000276256 | 1.456334e-01 | 1.587725e-01 | 1.663422e-01 | -7.569715e-03 | 9.946371e-01 | 1.000000e+00 | 0.022176195 | 0.016113771 | 0.023395954 | ⋯ | 0.2252880825 | 0.3118314572 | 0.3236612085 | -1.182975e-02 | 6.799064e-01 | 0.849895931 | NA | NA | NA | NA |
| ENSG00000278384.ENSG00000278384 | 8.811104e-03 | 1.040108e-02 | 1.052278e-02 | -1.217001e-04 | 5.977625e-01 | 9.991189e-01 | 0.003029215 | 0.001449831 | 0.003199408 | ⋯ | 0.0087212489 | 0.0138309448 | 0.0128132871 | 1.017658e-03 | 1.548784e-01 | 0.590868757 | NA | NA | NA | NA |
| ENSG00000273748.ENSG00000273748 | 8.629686e-02 | 9.038450e-02 | 8.604669e-02 | 4.337806e-03 | 4.265947e-01 | 8.149871e-01 | 0.068696288 | 0.080482907 | 0.072066698 | ⋯ | 0.0876646108 | 0.1369676261 | 0.1285271979 | 8.440428e-03 | 2.004931e-01 | 0.646712417 | NA | NA | NA | NA |
| ENSG00000271254.ENSG00000271254 | 1.367201e-02 | 1.176190e-02 | 1.199557e-02 | -2.336686e-04 | 1.048407e-01 | 2.843984e-01 | 0.005279681 | 0.004404255 | 0.005575911 | ⋯ | 0.0095123359 | 0.0147881250 | 0.0139754977 | 8.126273e-04 | 2.285474e-01 | 0.674288547 | NA | NA | NA | NA |
| ENSG00000276345.ENSG00000276345 | 2.556739e-02 | 1.401183e-02 | 1.560030e-02 | -1.588466e-03 | 8.243541e-01 | 1.000000e+00 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000276017.ENSG00000276017 | 3.954335e-05 | 9.976801e-06 | 2.048383e-05 | -1.050703e-05 | 9.858771e-01 | 1.000000e+00 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
| ENSG00000278817.ENSG00000278817 | 1.109546e-02 | 8.310894e-03 | 8.641444e-03 | -3.305493e-04 | 7.548021e-03 | 2.987236e-02 | 0.013793552 | 0.015746266 | 0.014561158 | ⋯ | 0.0024667705 | 0.0030916251 | 0.0036242592 | -5.326341e-04 | 9.699135e-01 | 1.000000000 | NA | NA | NA | NA |
| ENSG00000277196.ENSG00000277196 | 1.811506e-04 | 1.545324e-04 | 1.071847e-04 | 4.734772e-05 | 5.457040e-08 | 4.813223e-07 | 0.000000000 | 0.000000000 | 0.000000000 | ⋯ | 0.0000000000 | 0.0000000000 | 0.0000000000 | 0.000000e+00 | NaN | NaN | NA | NA | NA | NA |
options(repr.plot.width=10, repr.plot.height=10)
ggplot(mgv, aes(x=mean, y=bio, colour=subpopshort)) +
geom_point(size=5)
ggplot(mgv, aes(x=subpopshort, y=bio, colour=subpopshort)) +
geom_boxplot() +
theme(axis.text.x = element_text(angle = 45, hjust=1))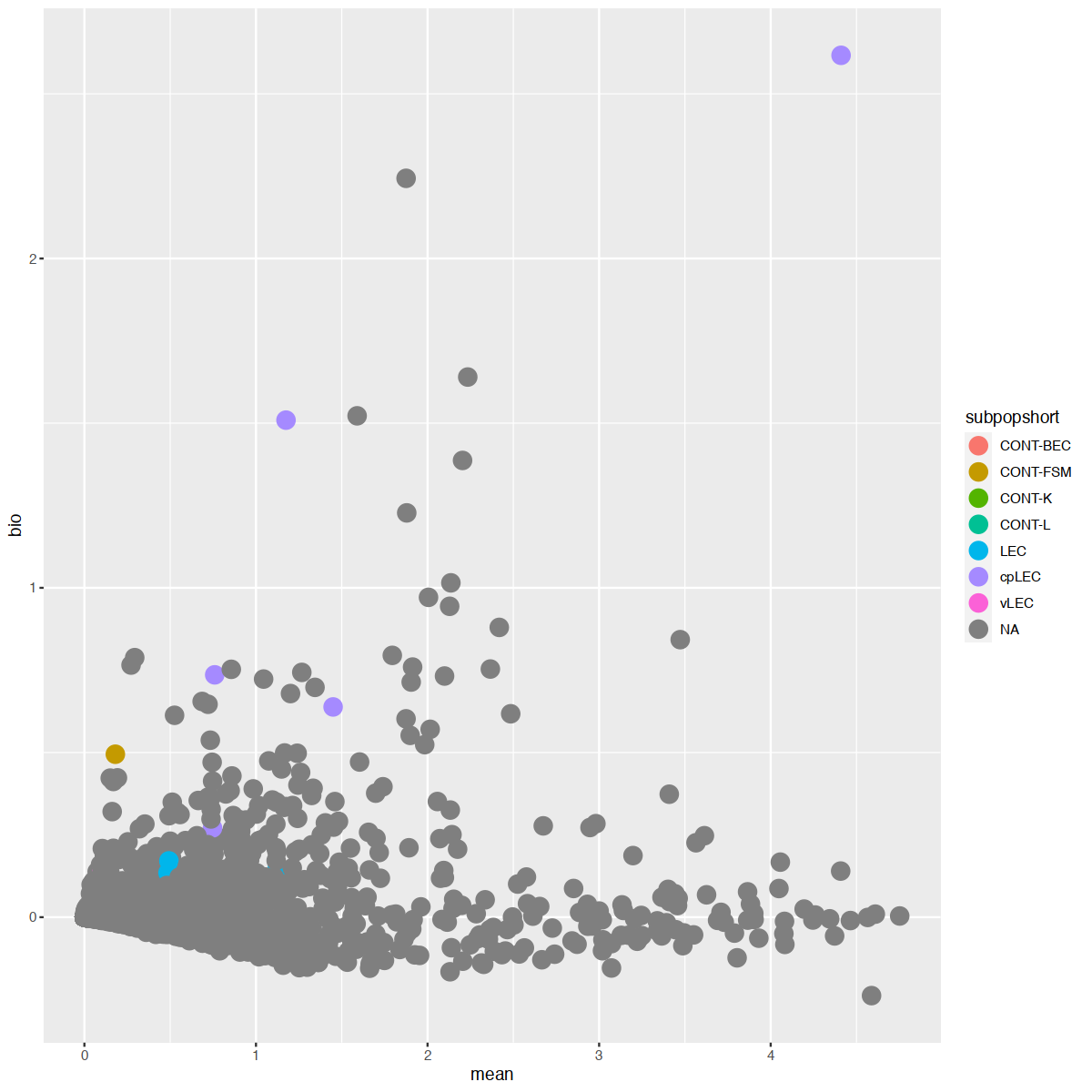
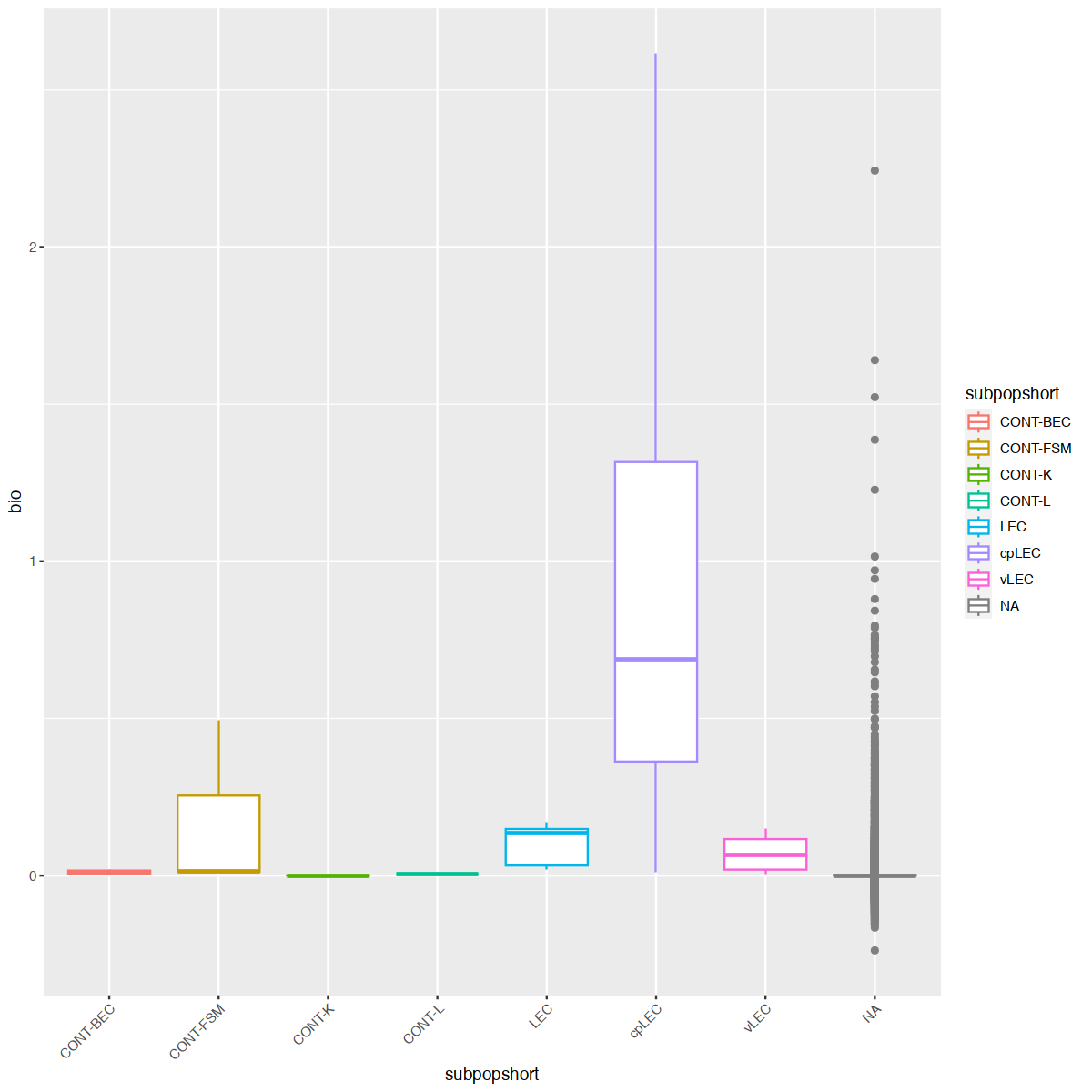
nm <- names(per_block)
names(nm) <- nm
nm- donor1_fat
- 'donor1_fat'
- donor1_skin
- 'donor1_skin'
- donor2_fat
- 'donor2_fat'
- donor3_fat
- 'donor3_fat'
- donor3_skin
- 'donor3_skin'
- donor4_fat
- 'donor4_fat'
- donor4_skin
- 'donor4_skin'
- donor5_recovery
- 'donor5_recovery'
- donor6_fat
- 'donor6_fat'
- donor6_skin
- 'donor6_skin'
- donor7_fat
- 'donor7_fat'
- donor7_skin
- 'donor7_skin'
options(repr.plot.width=6, repr.plot.height=6)
per_block_ggs <- lapply(nm, function(u) {
z <- per_block[[u]] %>% as.data.frame %>% rownames_to_column("gene_name") %>%
left_join(markers, multiple = "first")
p <- ggplot(z, aes(x=subpopshort, y=-log10(FDR), colour=subpopshort)) +
geom_boxplot() +
theme(axis.text.x = element_text(angle = 45, hjust=1)) +
ggtitle(u)
print(p)
})Joining with `by = join_by(gene_name)`
Warning message:
“Removed 5932 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 1984 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 2469 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 1555 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 3137 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 3732 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 2029 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 1699 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 2766 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 3088 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 2705 rows containing non-finite values (`stat_boxplot()`).”
Joining with `by = join_by(gene_name)`
Warning message:
“Removed 2229 rows containing non-finite values (`stat_boxplot()`).”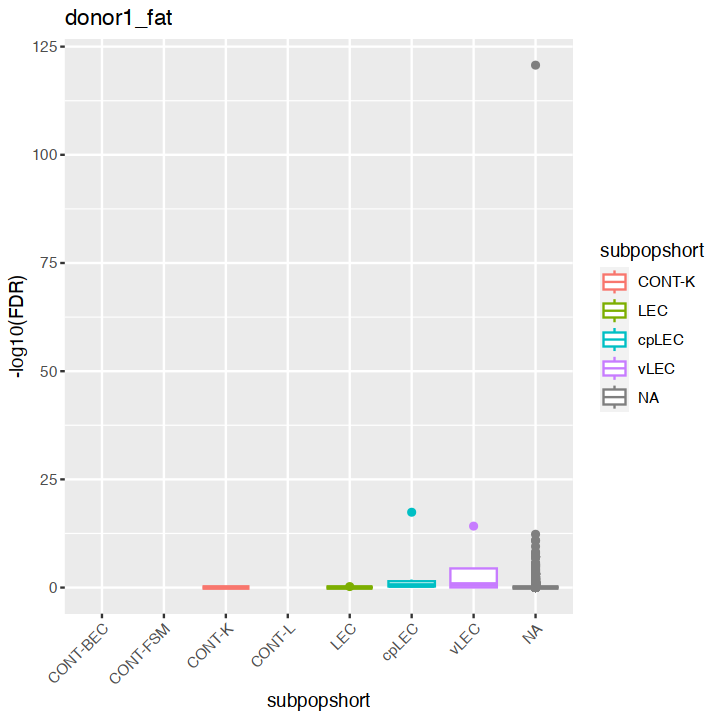
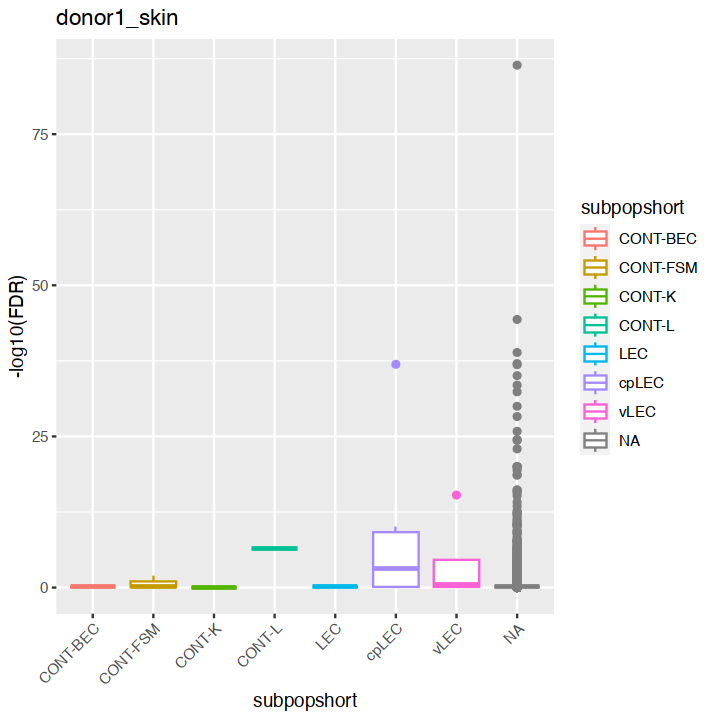
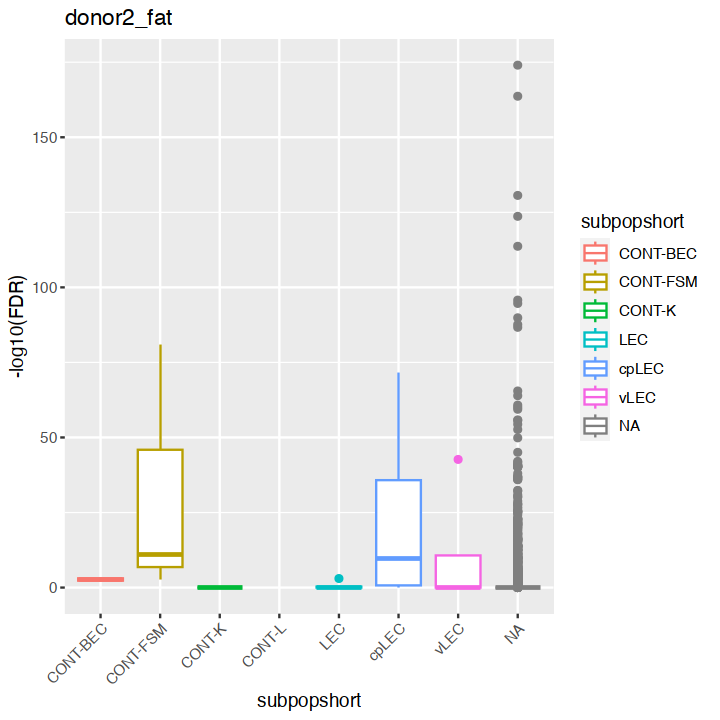
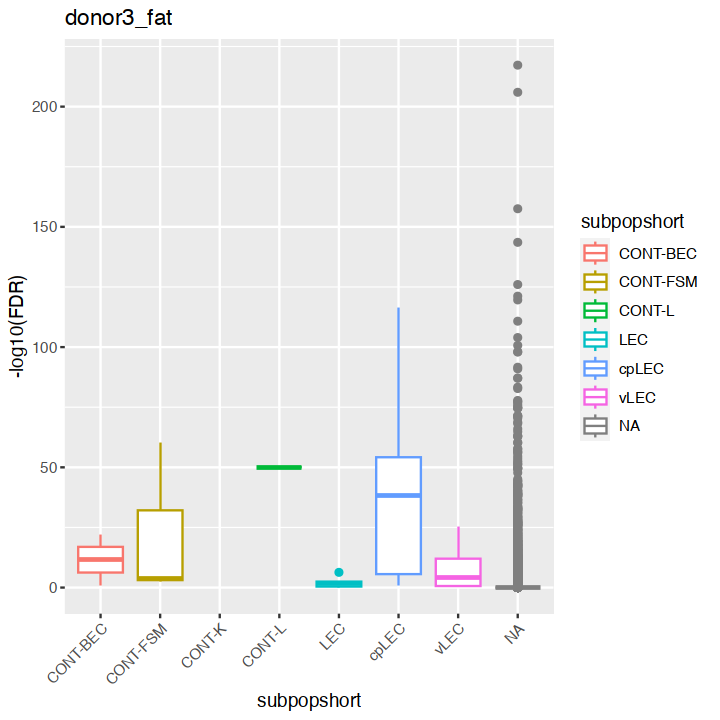
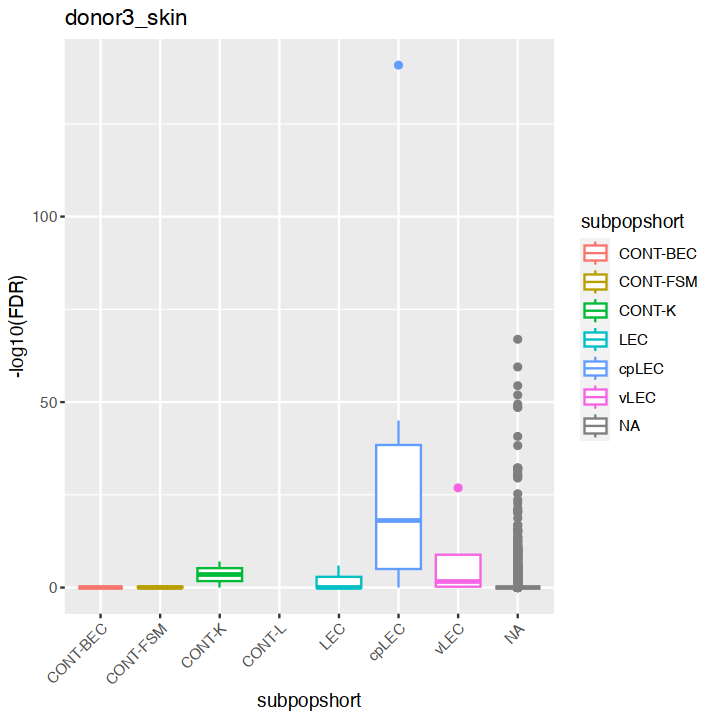
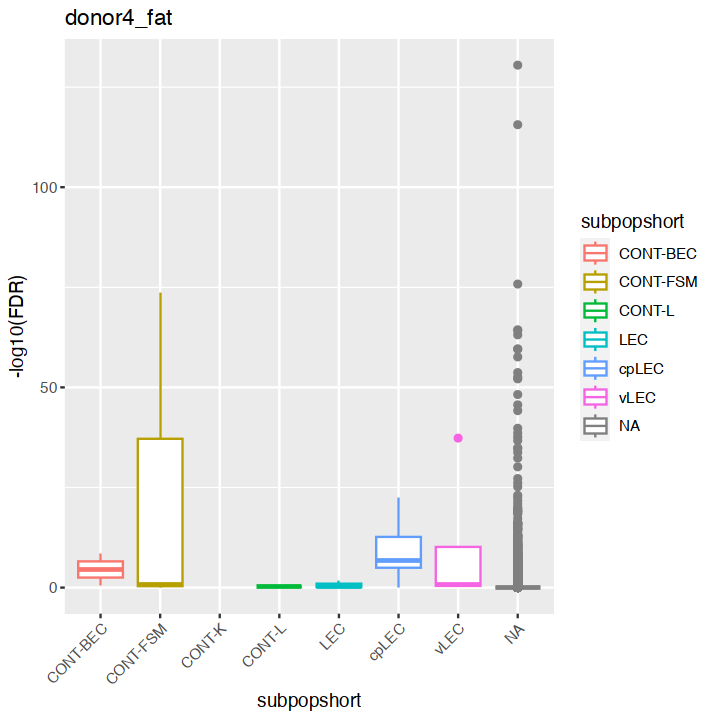
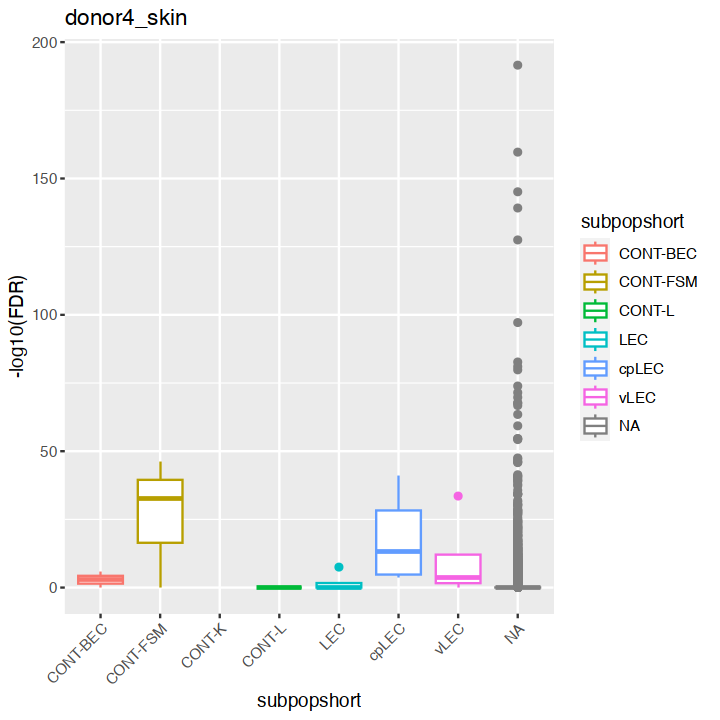
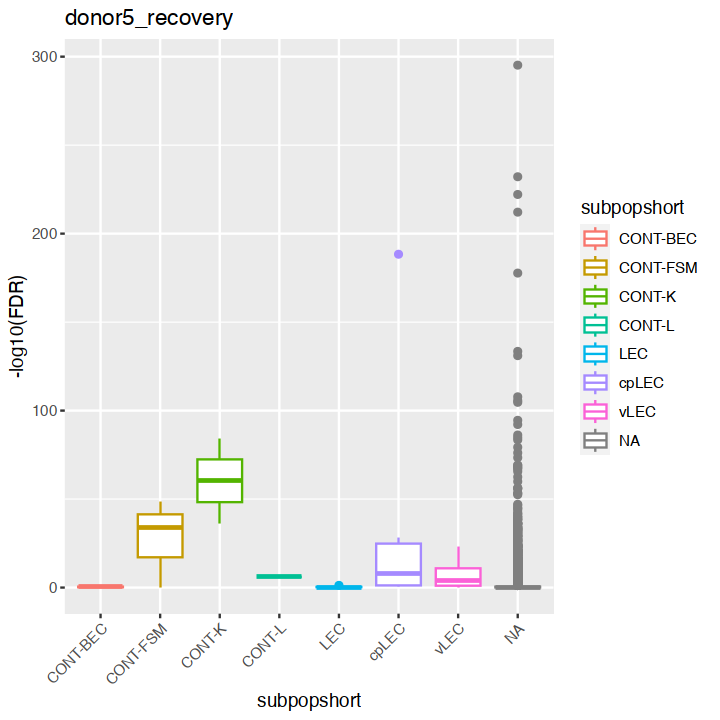
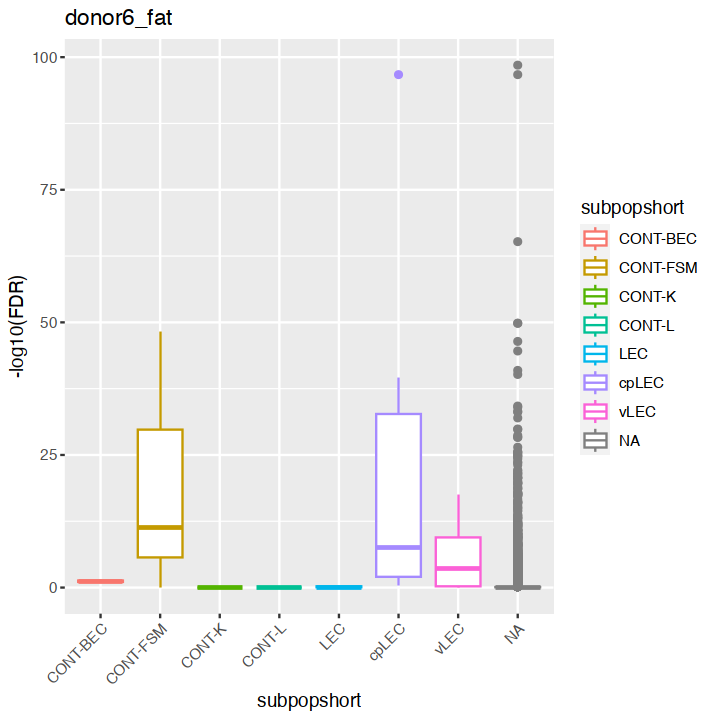
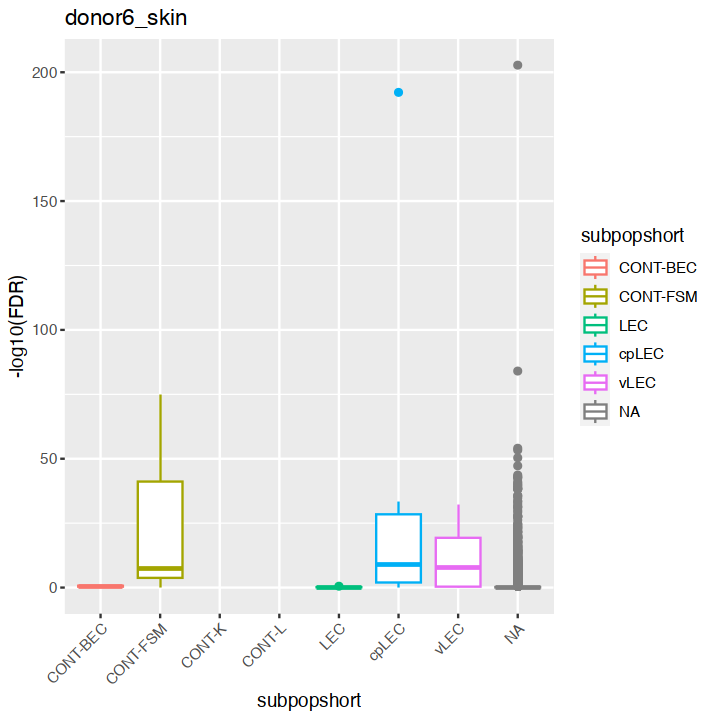
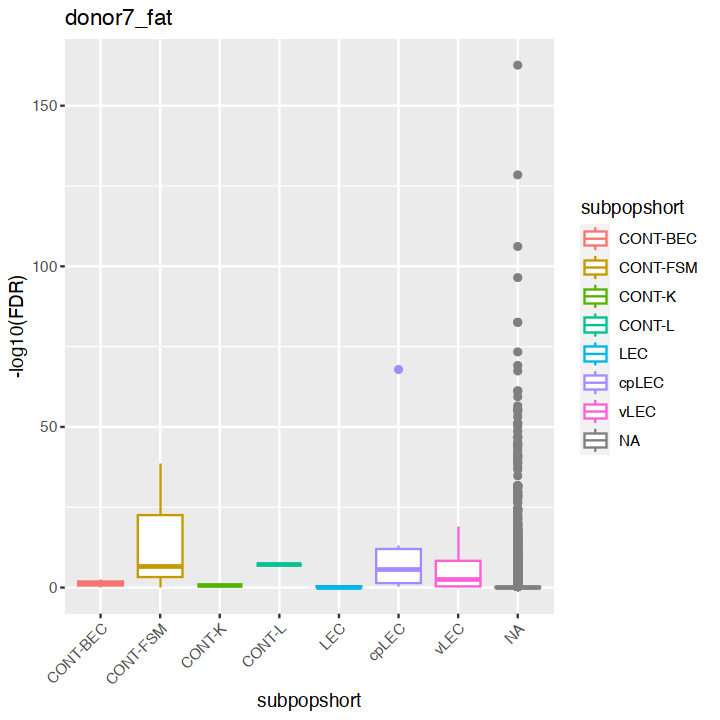
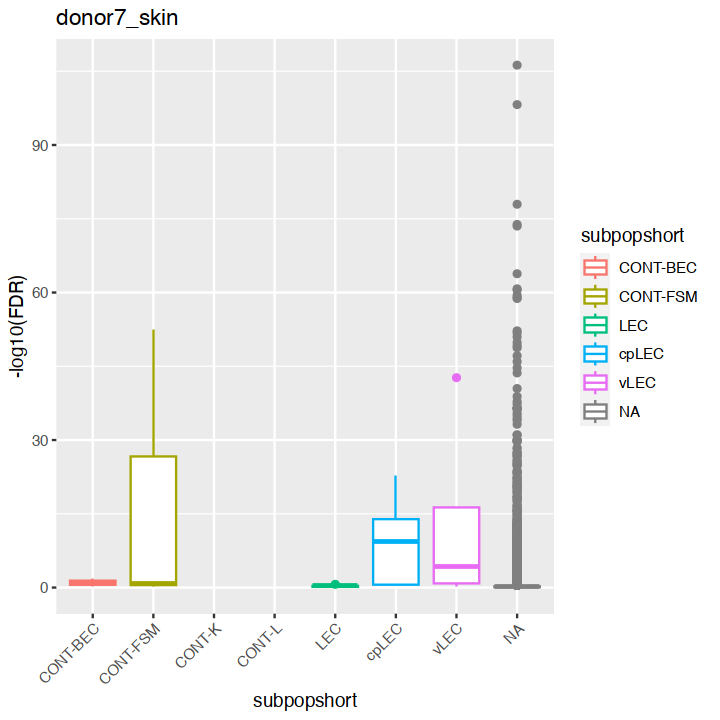
options(repr.plot.width=20, repr.plot.height=20)
ggarrange(plotlist = per_block_ggs, nrow = 3, ncol = 4)Warning message:
“Removed 5932 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 1984 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 2469 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 1555 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 3137 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 3732 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 2029 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 1699 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 2766 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 3088 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 2705 rows containing non-finite values (`stat_boxplot()`).”
Warning message:
“Removed 2229 rows containing non-finite values (`stat_boxplot()`).”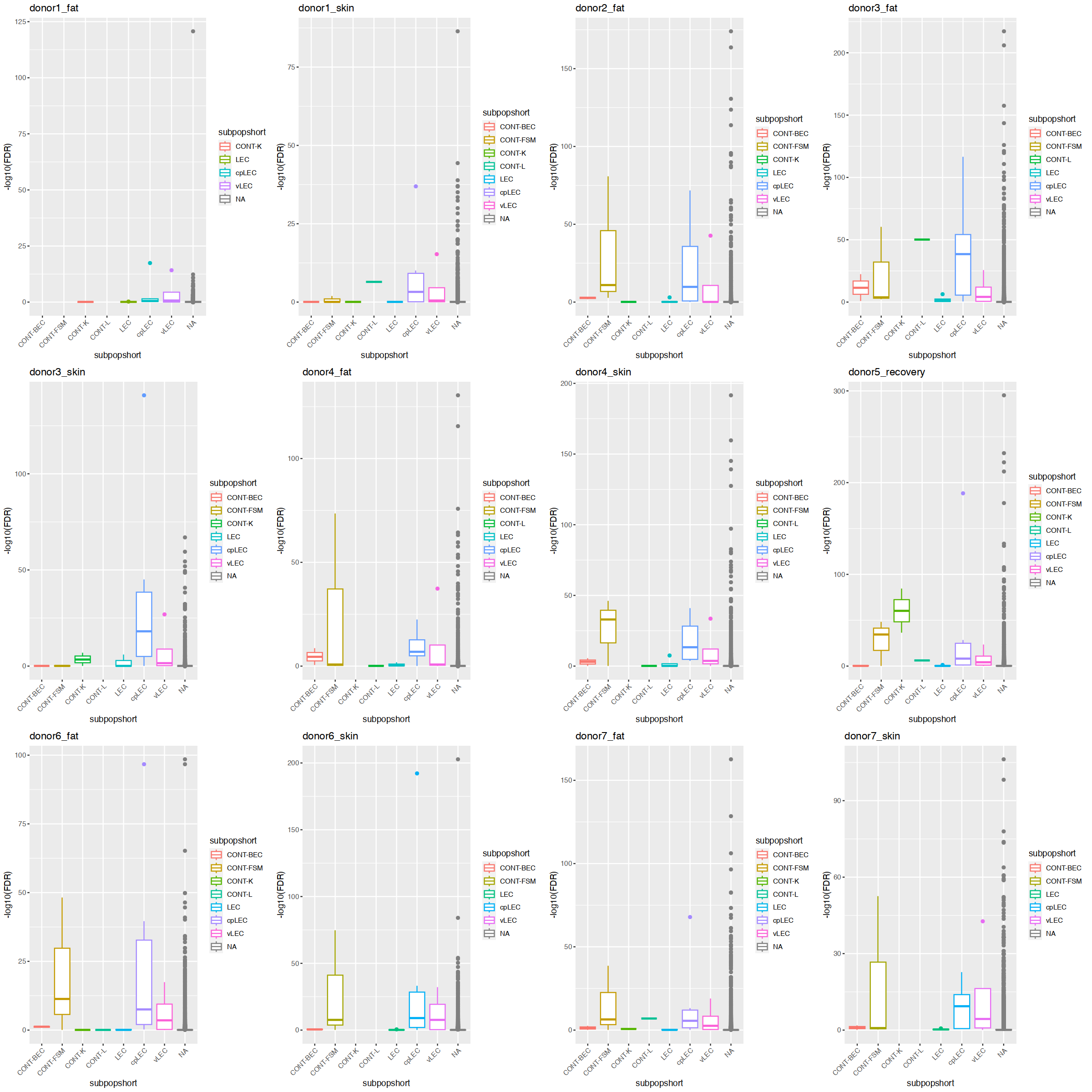
uniq_markers <- unique(markers$gene_name)
uniq_markers- 'ENSG00000117707.PROX1'
- 'ENSG00000162493.PDPN'
- 'ENSG00000133800.LYVE1'
- 'ENSG00000137077.CCL21'
- 'ENSG00000189056.RELN'
- 'ENSG00000129048.ACKR4'
- 'ENSG00000108691.CCL2'
- 'ENSG00000081041.CXCL2'
- 'ENSG00000013297.CLDN11'
- 'ENSG00000272398.CD24'
- 'ENSG00000149564.ESAM'
- 'ENSG00000170017.ALCAM'
- 'ENSG00000108821.COL1A1'
- 'ENSG00000113721.PDGFRB'
- 'ENSG00000107796.ACTA2'
- 'ENSG00000186081.KRT5'
- 'ENSG00000186847.KRT14'
- 'ENSG00000081237.PTPRC'
- 'ENSG00000102755.FLT1'
- 'ENSG00000154721.JAM2'
- 'ENSG00000090339.ICAM1'
- 'ENSG00000103335.PIEZO1'
- 'ENSG00000154864.PIEZO2'
per_block_umaps <- lapply(nm, function(u) {
s <- sce[,sce$Sample==u]
mgv <- modelGeneVar(s)
hvgs <- mgv %>% as.data.frame %>% rownames_to_column("gene_name") %>%
arrange(p.value) %>% head(500) %>% pull("gene_name")
s <- runPCA(s, subset_row=hvgs)
s <- runUMAP(s)
exp <- as.matrix(logcounts(s)[uniq_markers,])
data.frame(
rowname = rownames(colData(s)), Sample = s$Sample,
UMAP = reducedDim(s, "UMAP"), t(exp)
) %>% return()
})Warning message in (function (A, nv = 5, nu = nv, maxit = 1000, work = nv + 7, reorth = TRUE, :
“You're computing too large a percentage of total singular values, use a standard svd instead.”pdf("figures/umaps-markers-across-samples.pdf", width=10, height=8)
for(i in seq_len(length(uniq_markers))) {
m <- markers %>% dplyr::filter(gene_name==uniq_markers[i]) %>% pull("subpopulation")
print(ggplot(bind_rows(per_block_umaps), aes_string(x="UMAP.1", y="UMAP.2", colour = uniq_markers[i])) +
geom_point() +
scale_colour_gradient(low = "grey80", high = "blue") +
facet_wrap(~Sample, scales = "free") +
theme(legend.position="top") +
ggtitle(m)
)
}
dev.off()Warning message:
“`aes_string()` was deprecated in ggplot2 3.0.0.
ℹ Please use tidy evaluation idioms with `aes()`.
ℹ See also `vignette("ggplot2-in-packages")` for more information.”
pdf: 2
ms <- bind_rows(per_block_umaps) %>% melt(id.vars = c("rowname", "Sample", "UMAP.1", "UMAP.2")) %>% split("Sample")
ms
$Sample =
| rowname | Sample | UMAP.1 | UMAP.2 | variable | value | |
|---|---|---|---|---|---|---|
| <chr> | <chr> | <dbl> | <dbl> | <fct> | <dbl> | |
| 1 | donor1_fat.AAAGAACCACATACGT-1 | donor1_fat | -2.0484646 | -1.1637262 | ENSG00000117707.PROX1 | 1.1180317 |
| 2 | donor1_fat.AAAGGTAGTCCGGTCA-1 | donor1_fat | 2.8242526 | -1.0391492 | ENSG00000117707.PROX1 | 0.0000000 |
| 3 | donor1_fat.AAAGGTATCTCTCTTC-1 | donor1_fat | 0.9381566 | 0.6268698 | ENSG00000117707.PROX1 | 1.0517659 |
| 4 | donor1_fat.AACCACACAGTGGCTC-1 | donor1_fat | -0.6080225 | -0.6364202 | ENSG00000117707.PROX1 | 0.0000000 |
| 5 | donor1_fat.AACGGGATCTGCTGAA-1 | donor1_fat | -1.0524757 | -1.6120028 | ENSG00000117707.PROX1 | 0.0000000 |
| 6 | donor1_fat.AAGAACATCTGTGCAA-1 | donor1_fat | 1.5718066 | 0.7489513 | ENSG00000117707.PROX1 | 2.0521759 |
| 7 | donor1_fat.AAGCATCTCGATCCCT-1 | donor1_fat | 1.7909548 | 0.8797356 | ENSG00000117707.PROX1 | 1.7869727 |
| 8 | donor1_fat.AAGCGAGAGTATCCTG-1 | donor1_fat | -1.0558064 | 1.0522912 | ENSG00000117707.PROX1 | 2.0656439 |
| 9 | donor1_fat.AAGTTCGAGGCAGGTT-1 | donor1_fat | -1.0539844 | -0.7540492 | ENSG00000117707.PROX1 | 0.8052045 |
| 10 | donor1_fat.AATCGACCAATTGCAC-1 | donor1_fat | -1.3530891 | -1.1469282 | ENSG00000117707.PROX1 | 2.2499633 |
| 11 | donor1_fat.ACAAAGACAATGACCT-1 | donor1_fat | 1.8577221 | 0.7200912 | ENSG00000117707.PROX1 | 0.7807234 |
| 12 | donor1_fat.ACAAAGACACTAACGT-1 | donor1_fat | 1.6504609 | -0.9285525 | ENSG00000117707.PROX1 | 0.0000000 |
| 13 | donor1_fat.ACAACCAAGGGCTTCC-1 | donor1_fat | -0.6354316 | 1.3237580 | ENSG00000117707.PROX1 | 1.6079082 |
| 14 | donor1_fat.ACAGGGATCTCATAGG-1 | donor1_fat | -0.1048098 | 0.9759096 | ENSG00000117707.PROX1 | 2.5479505 |
| 15 | donor1_fat.ACATTTCCACTCCACT-1 | donor1_fat | 2.0356176 | -0.3929195 | ENSG00000117707.PROX1 | 0.0000000 |
| 16 | donor1_fat.ACATTTCTCGATGGAG-1 | donor1_fat | 0.4631455 | 1.2451497 | ENSG00000117707.PROX1 | 0.0000000 |
| 17 | donor1_fat.ACCCTTGTCACCATAG-1 | donor1_fat | 0.8288960 | -0.8372497 | ENSG00000117707.PROX1 | 0.9320313 |
| 18 | donor1_fat.ACCTACCAGACCTCCG-1 | donor1_fat | 2.0905075 | 0.3380302 | ENSG00000117707.PROX1 | 0.0000000 |
| 19 | donor1_fat.ACGGAAGCACACCTAA-1 | donor1_fat | -3.9451654 | 0.2357324 | ENSG00000117707.PROX1 | 2.1732948 |
| 20 | donor1_fat.ACGTAGTTCCCGAAAT-1 | donor1_fat | 2.7871909 | 0.2391406 | ENSG00000117707.PROX1 | 0.0000000 |
| 21 | donor1_fat.ACGTTCCAGATGAATC-1 | donor1_fat | -3.5143168 | 0.4605635 | ENSG00000117707.PROX1 | 0.0000000 |
| 22 | donor1_fat.ACTCTCGAGCCGTTGC-1 | donor1_fat | -0.4711578 | -0.9177226 | ENSG00000117707.PROX1 | 0.0000000 |
| 23 | donor1_fat.ACTTCCGTCAGGAACG-1 | donor1_fat | -4.0332029 | 0.4111551 | ENSG00000117707.PROX1 | 2.6920234 |
| 24 | donor1_fat.AGACACTCAGGCGATA-1 | donor1_fat | 2.8228720 | -1.0549392 | ENSG00000117707.PROX1 | 0.0000000 |
| 25 | donor1_fat.AGACCATTCAAAGGTA-1 | donor1_fat | -0.2053631 | 0.8217293 | ENSG00000117707.PROX1 | 1.4683445 |
| 26 | donor1_fat.AGAGAGCCAAGCCCAC-1 | donor1_fat | 2.1387318 | -0.4795322 | ENSG00000117707.PROX1 | 0.0000000 |
| 27 | donor1_fat.AGAGCCCAGTCTAGCT-1 | donor1_fat | -0.9772060 | -1.4272784 | ENSG00000117707.PROX1 | 0.9130940 |
| 28 | donor1_fat.AGATCCAGTGCAGTGA-1 | donor1_fat | -1.1057518 | -1.5379166 | ENSG00000117707.PROX1 | 2.6862529 |
| 29 | donor1_fat.AGATCGTGTTGCGTAT-1 | donor1_fat | 0.3803560 | 0.7960730 | ENSG00000117707.PROX1 | 1.5156637 |
| 30 | donor1_fat.AGGAGGTCAATACGAA-1 | donor1_fat | -0.5325592 | -1.5191154 | ENSG00000117707.PROX1 | 0.0000000 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| 512273 | donor7_skin.TTTATGCTCGTGTCAA-1 | donor7_skin | 0.44066022 | -2.3981223 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512274 | donor7_skin.TTTCACAAGCGATGGT-1 | donor7_skin | -2.32789935 | 0.5459931 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512275 | donor7_skin.TTTCACAGTTGTAGCT-1 | donor7_skin | -0.63556482 | -1.3933723 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512276 | donor7_skin.TTTCAGTTCAATCGGT-1 | donor7_skin | -0.62373890 | -1.2646084 | ENSG00000154864.PIEZO2 | 0.7371799 |
| 512277 | donor7_skin.TTTCATGGTACAGTCT-1 | donor7_skin | 1.56131671 | 2.2107785 | ENSG00000154864.PIEZO2 | 1.0879936 |
| 512278 | donor7_skin.TTTCATGGTGGCTGCT-1 | donor7_skin | 1.27958320 | 1.3705374 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512279 | donor7_skin.TTTCATGTCCTTATGT-1 | donor7_skin | 1.51498244 | -2.0856054 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512280 | donor7_skin.TTTCCTCAGCGGACAT-1 | donor7_skin | -1.95951611 | 0.0865000 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512281 | donor7_skin.TTTCCTCCACTCTAGA-1 | donor7_skin | 3.84445618 | 0.9458799 | ENSG00000154864.PIEZO2 | 1.4716415 |
| 512282 | donor7_skin.TTTCCTCGTACAAAGT-1 | donor7_skin | -1.86466755 | 0.1249833 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512283 | donor7_skin.TTTCCTCGTAGCGCTC-1 | donor7_skin | 0.96789597 | -1.2941531 | ENSG00000154864.PIEZO2 | 1.0866399 |
| 512284 | donor7_skin.TTTCCTCGTGGCCCAT-1 | donor7_skin | -1.77742146 | 1.0157657 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512285 | donor7_skin.TTTCGATAGTGTCATC-1 | donor7_skin | 1.08103965 | -0.5591272 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512286 | donor7_skin.TTTCGATCAAGAAATC-1 | donor7_skin | -2.02880452 | -4.1955617 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512287 | donor7_skin.TTTCGATTCTGCCCTA-1 | donor7_skin | -2.95822276 | -1.6004554 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512288 | donor7_skin.TTTGACTCACCCTTAC-1 | donor7_skin | -1.19297369 | 0.3856518 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512289 | donor7_skin.TTTGACTCAGTAACGG-1 | donor7_skin | -2.75150790 | -0.7835479 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512290 | donor7_skin.TTTGACTGTTCTGACA-1 | donor7_skin | -1.31828757 | -0.2367129 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512291 | donor7_skin.TTTGATCCACATATGC-1 | donor7_skin | 0.71390794 | 2.2297013 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512292 | donor7_skin.TTTGATCCACTAGAGG-1 | donor7_skin | -1.83226659 | 0.5647108 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512293 | donor7_skin.TTTGATCGTCCCAAAT-1 | donor7_skin | 0.01913915 | 2.3483524 | ENSG00000154864.PIEZO2 | 2.3022276 |
| 512294 | donor7_skin.TTTGATCGTTCTCCAC-1 | donor7_skin | 0.83273528 | 2.6859512 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512295 | donor7_skin.TTTGATCTCTGTCTCG-1 | donor7_skin | -1.36970748 | -0.4502688 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512296 | donor7_skin.TTTGGAGAGCAAACAT-1 | donor7_skin | 2.42198656 | 0.8811707 | ENSG00000154864.PIEZO2 | 1.7446003 |
| 512297 | donor7_skin.TTTGGAGAGTACTCGT-1 | donor7_skin | -0.99745222 | 2.7946794 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512298 | donor7_skin.TTTGGAGGTACGTGTT-1 | donor7_skin | -0.40469517 | 3.2030871 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512299 | donor7_skin.TTTGGAGTCCATTTGT-1 | donor7_skin | -2.61639698 | -1.3488972 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512300 | donor7_skin.TTTGGAGTCTTACCGC-1 | donor7_skin | 0.74898480 | 2.0100686 | ENSG00000154864.PIEZO2 | 1.3394578 |
| 512301 | donor7_skin.TTTGGTTGTTGTAGCT-1 | donor7_skin | 0.60430000 | 2.4989591 | ENSG00000154864.PIEZO2 | 0.0000000 |
| 512302 | donor7_skin.TTTGTTGCAACTGTGT-1 | donor7_skin | -0.57576229 | -1.1000320 | ENSG00000154864.PIEZO2 | 2.3025932 |
pdf("figures/umaps-all-markers-each-sample.pdf", width=10, height=8)
for(i in seq_len(length(ms))) {
print(
ggplot(ms[[i]], aes(x=UMAP.1, y=UMAP.2, colour = value)) +
geom_point() +
scale_colour_gradient(low = "grey80", high = "blue") +
facet_wrap(~variable) +
theme(legend.position="top") +
ggtitle(names(ms)[i])
)
}
dev.off()
pdf: 2
d <- bind_rows(per_block_umaps)
d_rnormd <- d[,-c(1:4)] + rnorm(prod(dim(d)), mean = 0, sd = .001)
ds <- split(d_rnormd, d$Sample)pdf("figures/heatmaps-all-markers-each-sample.pdf", width=10, height=5)
for(i in seq_len(length(ds))) {
pheatmap(t(ds[[i]]), show_colnames = FALSE,
clustering_distance_cols = "correlation",
clustering_distance_rows = "correlation",
main = names(ds)[i]
)
}
dev.off()
pdf: 3
ds_norm <- ds
pdf("figures/heatmaps-all-markers-each-sample-normalized.pdf", width=10, height=5)
for(i in seq_len(length(ds))) {
qs <- apply(ds[[i]], 2, quantile, p=.90)
qs[qs < 1] <- 1
for(j in 1:ncol(ds[[i]]))
ds_norm[[i]][,j] <- ds_norm[[i]][,j]/qs[j]
ds_norm[[i]][ds_norm[[i]]>1] <- 1
pheatmap(t(ds_norm[[i]]), show_colnames = FALSE,
clustering_distance_cols = "correlation",
clustering_distance_rows = "correlation",
main = names(ds)[i]
)
}
dev.off()
pdf: 3
# contaminantsoptions(repr.plot.width=20, repr.plot.height=20)
ggplot(d, aes(
x = ENSG00000137077.CCL21,
y = ENSG00000108821.COL1A1)
) +
geom_jitter(width=.1, height = .5) +
facet_wrap(~Sample)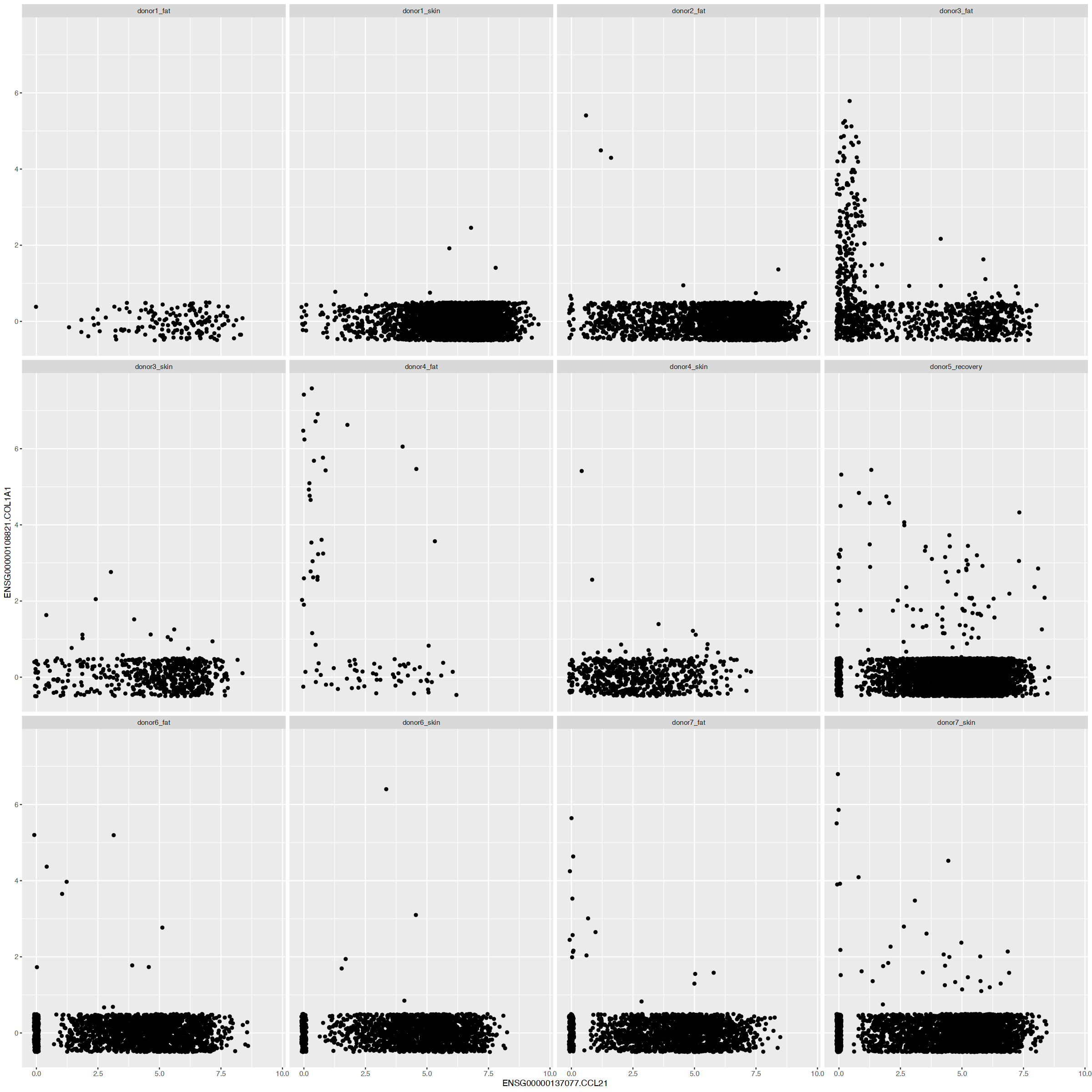
options(repr.plot.width=12, repr.plot.height=6)
boxplot(as.data.frame(d[,-(1:4)]), las=2)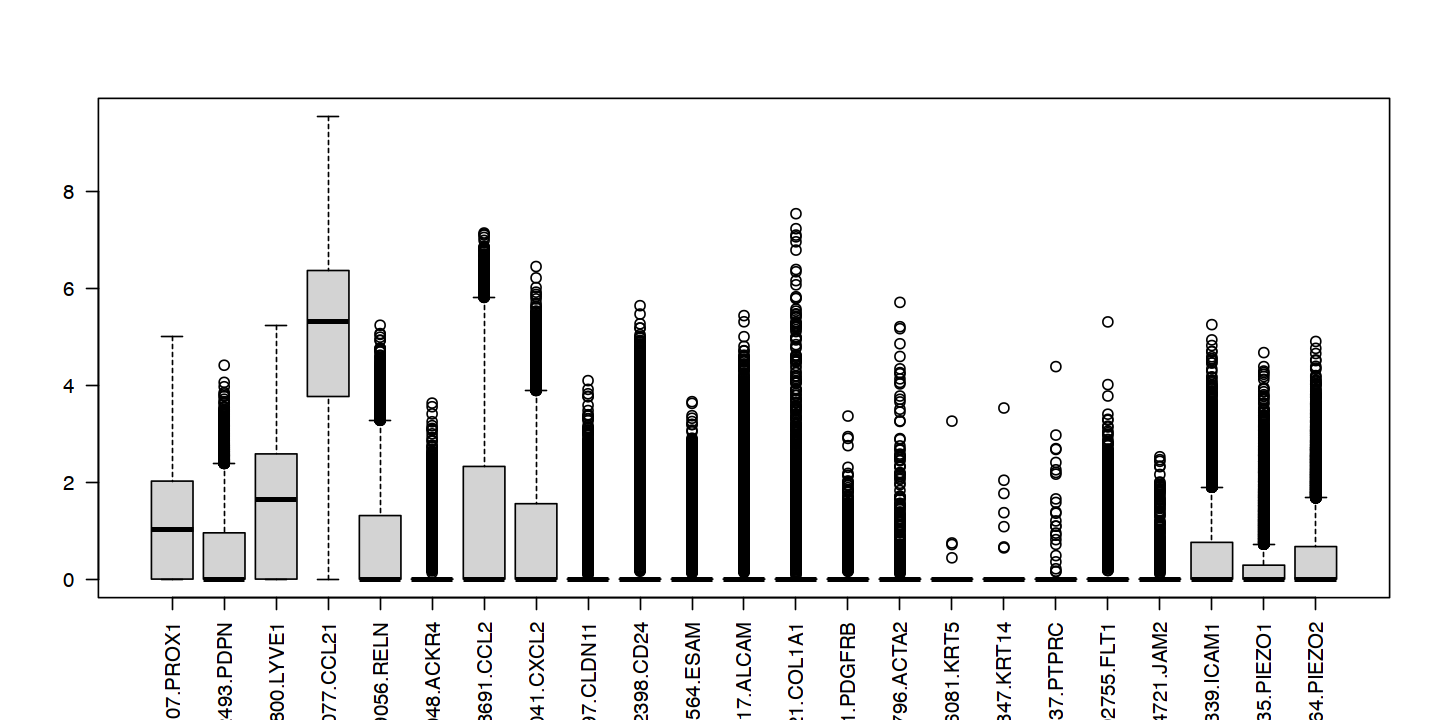
exp <- d[,-(1:4)] %>% as.matrix
p90 <- exp %>% apply(2, quantile, probs=.9)
p90- ENSG00000117707.PROX1
- 2.7040474964207
- ENSG00000162493.PDPN
- 1.69283581132588
- ENSG00000133800.LYVE1
- 3.26185058645611
- ENSG00000137077.CCL21
- 7.22691459652125
- ENSG00000189056.RELN
- 2.24996327230414
- ENSG00000129048.ACKR4
- 0
- ENSG00000108691.CCL2
- 3.71939503454669
- ENSG00000081041.CXCL2
- 2.89062607918324
- ENSG00000013297.CLDN11
- 0
- ENSG00000272398.CD24
- 0
- ENSG00000149564.ESAM
- 0.355130098838991
- ENSG00000170017.ALCAM
- 0
- ENSG00000108821.COL1A1
- 0
- ENSG00000113721.PDGFRB
- 0
- ENSG00000107796.ACTA2
- 0
- ENSG00000186081.KRT5
- 0
- ENSG00000186847.KRT14
- 0
- ENSG00000081237.PTPRC
- 0
- ENSG00000102755.FLT1
- 0
- ENSG00000154721.JAM2
- 0
- ENSG00000090339.ICAM1
- 1.88047655359566
- ENSG00000103335.PIEZO1
- 1.28664026201016
- ENSG00000154864.PIEZO2
- 1.73042322902796
col1a1 <- exp[,"ENSG00000108821.COL1A1"] / 2
col1a1[col1a1 > 1] <- 1
jam2 <- exp[,"ENSG00000154721.JAM2"] / 2
jam2[jam2 > 1] <- 1
prgfrb <- exp[,"ENSG00000113721.PDGFRB"] / 2
prgfrb[prgfrb > 1] <- 1
pdpn <- exp[,"ENSG00000162493.PDPN"] / p90["ENSG00000162493.PDPN"]
pdpn[pdpn > 1] <- 1
cxcl2 <- exp[,"ENSG00000081041.CXCL2"] / p90["ENSG00000081041.CXCL2"]
cxcl2[cxcl2 > 1] <- 1
ccl2 <- exp[,"ENSG00000108691.CCL2"] / p90["ENSG00000108691.CCL2"]
ccl2[ccl2 > 1] <- 1
ccl21 <- exp[,"ENSG00000137077.CCL21"] / p90["ENSG00000137077.CCL21"]
ccl21[ccl21 > 1] <- 1
lyve1 <- exp[,"ENSG00000133800.LYVE1"] / p90["ENSG00000133800.LYVE1"]
lyve1[lyve1 > 1] <- 1
prox1 <- exp[,"ENSG00000117707.PROX1"] / p90["ENSG00000117707.PROX1"]
prox1[prox1 > 1] <- 1
df <- data.frame(
Sample=d$Sample,
col1a1, jam2, prgfrb, ccl21, lyve1,
prox1, pdpn, ccl2, cxcl2
)
# noise? before filtering?
df[,-1] <- df[,-1] + rnorm(
nrow(df)*(ncol(df)-1),
mean=0, sd=.02
)
df| Sample | col1a1 | jam2 | prgfrb | ccl21 | lyve1 | prox1 | pdpn | ccl2 | cxcl2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| donor1_fat.AAAGAACCACATACGT-1 | donor1_fat | -6.177371e-03 | -0.0031744491 | 0.025607984 | 0.8637877 | 0.506185147 | 0.399910959 | -2.141122e-02 | -0.0147191045 | -0.0092179474 |
| donor1_fat.AAAGGTAGTCCGGTCA-1 | donor1_fat | -2.562640e-02 | 0.0135274094 | 0.016411063 | 0.6995753 | -0.030954370 | -0.020318782 | 1.272878e-02 | 0.0321146768 | -0.0155214929 |
| donor1_fat.AAAGGTATCTCTCTTC-1 | donor1_fat | 3.954838e-03 | -0.0047302147 | -0.058921807 | 1.0064774 | 0.009121395 | 0.403507005 | 9.707439e-01 | 0.0009019757 | -0.0097793427 |
| donor1_fat.AACCACACAGTGGCTC-1 | donor1_fat | -8.390417e-04 | -0.0049775072 | 0.004997854 | 0.9798234 | 0.630962393 | -0.022863151 | 3.719339e-01 | 0.6664521861 | 0.0467031244 |
| donor1_fat.AACGGGATCTGCTGAA-1 | donor1_fat | -3.294729e-02 | 0.0172184506 | 0.025801842 | 1.0099529 | 0.952636547 | 0.016039127 | 3.159890e-02 | -0.0161948350 | 0.0093164554 |
| donor1_fat.AAGAACATCTGTGCAA-1 | donor1_fat | -9.278279e-03 | -0.0432904238 | -0.004928558 | 0.6361269 | 0.021998822 | 0.764609556 | 7.688593e-03 | 0.4987814014 | 0.2286961444 |
| donor1_fat.AAGCATCTCGATCCCT-1 | donor1_fat | 8.961247e-03 | 0.0257253419 | -0.007706475 | 0.5688781 | 0.011359943 | 0.668748819 | -9.337751e-04 | -0.0031576763 | -0.0033508996 |
| donor1_fat.AAGCGAGAGTATCCTG-1 | donor1_fat | -2.025285e-02 | -0.0427843370 | 0.043382435 | 0.7318062 | 0.612130896 | 0.729646983 | 4.289729e-03 | 1.0105701492 | 0.5701520444 |
| donor1_fat.AAGTTCGAGGCAGGTT-1 | donor1_fat | -1.957310e-02 | -0.0155224185 | -0.014063269 | 0.7817754 | 0.948710864 | 0.299500667 | 7.823209e-01 | -0.0224818197 | -0.0198374980 |
| donor1_fat.AATCGACCAATTGCAC-1 | donor1_fat | 5.040912e-03 | -0.0047053116 | -0.001473258 | 0.6704721 | 0.547458265 | 0.829585304 | -8.579479e-05 | -0.0042205833 | -0.0122900824 |
| donor1_fat.ACAAAGACAATGACCT-1 | donor1_fat | -2.382496e-02 | 0.0412669633 | -0.004748681 | 0.7122134 | -0.008654046 | 0.306771378 | 4.814842e-01 | 0.3609760289 | 0.5507497536 |
| donor1_fat.ACAAAGACACTAACGT-1 | donor1_fat | -2.887057e-02 | -0.0007119733 | -0.011167208 | 0.2209485 | -0.004722751 | -0.006377754 | 1.007664e-02 | -0.0018593270 | 0.9934970903 |
| donor1_fat.ACAACCAAGGGCTTCC-1 | donor1_fat | 7.492564e-03 | 0.0063471223 | 0.009616054 | 0.7539768 | 0.873148301 | 0.580555478 | 5.203047e-01 | 0.3668938136 | -0.0005466140 |
| donor1_fat.ACAGGGATCTCATAGG-1 | donor1_fat | 2.690249e-02 | -0.0020851500 | 0.010444795 | 0.7800630 | 0.400894972 | 0.965109975 | -1.875359e-02 | 0.6930383825 | -0.0011001914 |
| donor1_fat.ACATTTCCACTCCACT-1 | donor1_fat | -2.288523e-02 | 0.0277930016 | -0.001592410 | 1.0105555 | 0.331541713 | -0.017978599 | 2.130233e-02 | 0.9701081869 | 0.6099380962 |
| donor1_fat.ACATTTCTCGATGGAG-1 | donor1_fat | 1.469623e-02 | 0.0315918627 | -0.030168437 | 0.7318597 | 0.807769612 | 0.013928482 | 2.950614e-02 | 0.8370519554 | 0.0040403227 |
| donor1_fat.ACCCTTGTCACCATAG-1 | donor1_fat | -2.389362e-02 | -0.0308304543 | 0.003702698 | 0.9223370 | 0.451392924 | 0.348397754 | 1.515303e-02 | 1.0203946074 | 0.6950916051 |
| donor1_fat.ACCTACCAGACCTCCG-1 | donor1_fat | -5.153461e-05 | -0.0112837207 | -0.016008569 | 1.0031394 | -0.006312337 | -0.006464281 | 3.518851e-03 | 1.0086026451 | -0.0328234172 |
| donor1_fat.ACGGAAGCACACCTAA-1 | donor1_fat | -4.068141e-02 | -0.0195332344 | -0.013722141 | 0.7616221 | 0.903429875 | 0.811012906 | -2.210629e-02 | 0.0126439890 | -0.0003801494 |
| donor1_fat.ACGTAGTTCCCGAAAT-1 | donor1_fat | 1.864060e-02 | 0.0445714489 | 0.006365741 | 0.8880584 | 0.704812734 | -0.033339004 | 1.545340e-03 | 0.5677575058 | -0.0066729671 |
| donor1_fat.ACGTTCCAGATGAATC-1 | donor1_fat | -2.603773e-02 | 0.0030491045 | 0.006140261 | 0.9205795 | 0.967926311 | 0.041954248 | 1.651663e-02 | -0.0126926382 | 0.0182637707 |
| donor1_fat.ACTCTCGAGCCGTTGC-1 | donor1_fat | -1.347052e-02 | -0.0266749866 | -0.005039600 | 0.8886155 | -0.016309032 | 0.003981775 | 4.496706e-02 | 0.0007821924 | -0.0156276358 |
| donor1_fat.ACTTCCGTCAGGAACG-1 | donor1_fat | 3.204020e-02 | -0.0264540226 | 0.001974596 | 0.5470836 | -0.025112726 | 1.016290121 | 8.666760e-01 | 0.0273618310 | -0.0189019862 |
| donor1_fat.AGACACTCAGGCGATA-1 | donor1_fat | -1.730093e-02 | 0.0059739468 | 0.015334581 | 0.5701491 | 0.476474856 | -0.016580222 | 9.025668e-01 | -0.0097566128 | 0.0045944003 |
| donor1_fat.AGACCATTCAAAGGTA-1 | donor1_fat | -1.537451e-02 | -0.0036618865 | 0.010825460 | 0.8017194 | 0.346596916 | 0.572483367 | 4.284791e-01 | 0.5251782663 | 0.6016392301 |
| donor1_fat.AGAGAGCCAAGCCCAC-1 | donor1_fat | -2.320340e-02 | -0.0068700812 | -0.003882135 | 0.9658855 | 0.574961429 | -0.007573551 | -2.751537e-02 | 1.0131715862 | -0.0037450447 |
| donor1_fat.AGAGCCCAGTCTAGCT-1 | donor1_fat | 9.452643e-03 | 0.0290292370 | -0.018340308 | 0.8366796 | 0.782972925 | 0.312903236 | 7.415997e-03 | -0.0047334983 | -0.0152212498 |
| donor1_fat.AGATCCAGTGCAGTGA-1 | donor1_fat | -1.201114e-02 | 0.0098563370 | 0.007610442 | 0.6463695 | 0.845765201 | 0.988111397 | -1.628989e-02 | -0.0451875636 | 0.4230879417 |
| donor1_fat.AGATCGTGTTGCGTAT-1 | donor1_fat | 8.615362e-03 | 0.0034245307 | 0.003521553 | 0.9543483 | 0.258806687 | 0.545596044 | 8.743513e-01 | 0.8007611046 | 0.0106937511 |
| donor1_fat.AGGAGGTCAATACGAA-1 | donor1_fat | 1.324703e-02 | -0.0081669716 | -0.002687446 | 1.0038940 | 0.705750068 | 0.024315192 | 2.940806e-03 | 0.0233627376 | 0.0038278500 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| donor7_skin.TTTATGCTCGTGTCAA-1 | donor7_skin | -0.0003990615 | 0.0296742640 | 0.0024537346 | -0.0022613577 | 0.005936938 | -0.0152438542 | 0.011365044 | 0.0266668524 | 0.993266362 |
| donor7_skin.TTTCACAAGCGATGGT-1 | donor7_skin | 0.0724061217 | 0.0163329879 | -0.0121939738 | 0.8780212054 | 0.915039457 | -0.0022887873 | -0.003619139 | 0.6576163700 | -0.022271624 |
| donor7_skin.TTTCACAGTTGTAGCT-1 | donor7_skin | -0.0193141786 | 0.0291989017 | 0.0173694292 | 0.6724375849 | -0.014048362 | 0.8438412083 | -0.014767578 | 0.3371750161 | 1.012992834 |
| donor7_skin.TTTCAGTTCAATCGGT-1 | donor7_skin | -0.0089066053 | 0.0535302078 | 0.0057074223 | 0.9399958719 | 0.837298673 | 0.7853596621 | 0.001170393 | 0.5677482638 | -0.015832060 |
| donor7_skin.TTTCATGGTACAGTCT-1 | donor7_skin | 0.0168256604 | -0.0141936968 | 0.0342388298 | 0.8215068327 | 0.956895869 | 0.9802217122 | -0.014461001 | -0.0221556456 | 0.024801192 |
| donor7_skin.TTTCATGGTGGCTGCT-1 | donor7_skin | -0.0070698679 | 0.0068699903 | -0.0091017995 | 0.2931002673 | 0.810785461 | 1.0026773554 | 0.492932180 | 0.0548301164 | -0.010028765 |
| donor7_skin.TTTCATGTCCTTATGT-1 | donor7_skin | -0.0244871095 | 0.0021237005 | 0.0569417374 | 0.2855903543 | 0.008329916 | 0.7890122466 | 0.667711838 | 0.8982960297 | 0.859786864 |
| donor7_skin.TTTCCTCAGCGGACAT-1 | donor7_skin | 0.0101579819 | -0.0072413576 | -0.0085471731 | 0.9655089864 | 0.537852124 | -0.0006230239 | 0.019402825 | 0.9072832572 | 0.883812935 |
| donor7_skin.TTTCCTCCACTCTAGA-1 | donor7_skin | 0.0078304239 | -0.0350495412 | -0.0009815983 | 0.4845271662 | 0.990405026 | 0.8220684204 | 0.019326832 | 0.0022491603 | -0.011941081 |
| donor7_skin.TTTCCTCGTACAAAGT-1 | donor7_skin | -0.0362198769 | -0.0103053609 | 0.0227414740 | 0.8186186769 | 0.576536763 | 0.9798925477 | 0.006112943 | 0.0073708300 | -0.020194533 |
| donor7_skin.TTTCCTCGTAGCGCTC-1 | donor7_skin | -0.0272635933 | -0.0507201323 | 0.0359429627 | 0.3640793752 | 0.693701713 | 0.2074575138 | 0.383341644 | 0.5185157652 | 0.002529597 |
| donor7_skin.TTTCCTCGTGGCCCAT-1 | donor7_skin | -0.0141176115 | 0.0140864067 | 0.0219909984 | 0.5788112613 | 0.027949546 | 0.0088000969 | -0.032908415 | 0.6754636217 | 1.001629578 |
| donor7_skin.TTTCGATAGTGTCATC-1 | donor7_skin | 0.0094453692 | -0.0021904036 | 0.0250803613 | 0.7851348844 | 0.960259296 | 0.3687019867 | -0.026586277 | 0.0046595506 | 0.021779652 |
| donor7_skin.TTTCGATCAAGAAATC-1 | donor7_skin | -0.0242377823 | -0.0403578822 | -0.0062238060 | -0.0191509953 | -0.003257678 | 0.9991128536 | -0.008962441 | 0.0064038129 | -0.021909010 |
| donor7_skin.TTTCGATTCTGCCCTA-1 | donor7_skin | -0.0032107839 | 0.0155023708 | 0.0389190531 | -0.0136244979 | 0.020727457 | 0.7525883420 | 0.782999736 | 0.0112400347 | 0.463305575 |
| donor7_skin.TTTGACTCACCCTTAC-1 | donor7_skin | -0.0001306085 | -0.0178902702 | 0.0078969840 | 0.9241689730 | 0.547651788 | -0.0120514165 | 1.009696181 | 0.0331389587 | -0.046051959 |
| donor7_skin.TTTGACTCAGTAACGG-1 | donor7_skin | 0.0050997361 | -0.0118696652 | 0.0130506358 | 0.5997844688 | 0.544050749 | -0.0387092503 | -0.021861516 | 1.0054835865 | 0.617677334 |
| donor7_skin.TTTGACTGTTCTGACA-1 | donor7_skin | -0.0033006433 | -0.0167376666 | -0.0095107318 | 0.7667512684 | 0.013864044 | 0.5748190268 | 0.007200415 | 0.8863525341 | -0.012955067 |
| donor7_skin.TTTGATCCACATATGC-1 | donor7_skin | 0.0086986293 | 0.0001716679 | -0.0063615097 | 0.7438463843 | -0.017501750 | 0.0030111330 | 0.043734232 | 0.4958974983 | 0.012600984 |
| donor7_skin.TTTGATCCACTAGAGG-1 | donor7_skin | -0.0032251796 | -0.0021914976 | 0.0128733049 | 1.0157908962 | -0.001787367 | -0.0052518524 | -0.008076685 | -0.0090272773 | 0.973062128 |
| donor7_skin.TTTGATCGTCCCAAAT-1 | donor7_skin | -0.0195745681 | 0.0250537166 | -0.0088243499 | -0.0039134482 | -0.022815206 | -0.0144200855 | 0.005719765 | 0.0196336264 | 0.735776560 |
| donor7_skin.TTTGATCGTTCTCCAC-1 | donor7_skin | -0.0434723500 | -0.0252945210 | 0.0179085736 | 0.6929848846 | 0.962433259 | 0.7529268707 | 0.020575347 | 0.0015099153 | 0.024224592 |
| donor7_skin.TTTGATCTCTGTCTCG-1 | donor7_skin | 0.0070041822 | -0.0115678572 | 0.0243900636 | 0.5309536652 | 0.608532916 | -0.0109624939 | 0.828145276 | 0.3868799389 | 0.945091813 |
| donor7_skin.TTTGGAGAGCAAACAT-1 | donor7_skin | 0.0041887005 | 0.0139418241 | -0.0088589254 | 0.6603070090 | 0.998915741 | 0.8172057627 | -0.022135229 | -0.0187547122 | 0.979596396 |
| donor7_skin.TTTGGAGAGTACTCGT-1 | donor7_skin | 0.0054559858 | 0.0175492243 | 0.0156962339 | 0.7827001774 | 0.987185754 | 0.6964182957 | 0.036893497 | -0.0010041689 | -0.004765871 |
| donor7_skin.TTTGGAGGTACGTGTT-1 | donor7_skin | 0.0190830933 | 0.0145798248 | -0.0356252672 | 1.0102405349 | 0.542353546 | 0.0361041893 | -0.022489221 | -0.0096067625 | 0.018684895 |
| donor7_skin.TTTGGAGTCCATTTGT-1 | donor7_skin | -0.0027773493 | -0.0189338551 | 0.0359169146 | 0.0009260175 | -0.000390323 | 0.4425000838 | -0.012844999 | 0.5196031753 | 0.438295525 |
| donor7_skin.TTTGGAGTCTTACCGC-1 | donor7_skin | 0.0331070097 | -0.0074154506 | 0.0028981773 | 0.4783320616 | 0.747969470 | 0.4664506242 | 0.001862201 | -0.0004225433 | 0.007729157 |
| donor7_skin.TTTGGTTGTTGTAGCT-1 | donor7_skin | -0.0246628673 | -0.0032276620 | -0.0063759478 | 0.8240156451 | 1.017997709 | 0.0117801748 | 0.006195800 | 0.0052499922 | 0.016706311 |
| donor7_skin.TTTGTTGCAACTGTGT-1 | donor7_skin | 0.0128099107 | 0.0109123962 | -0.0250518974 | 0.8200733693 | 0.914873636 | 0.6920174360 | -0.027329639 | 0.3451530485 | 0.010775754 |
df$keep <- df %>% with((col1a1+jam2+prgfrb) < .1)
mean(!df$keep)
sum(!df$keep)
with(df, table(Sample, keep))
0.0311574032504265
694
keep
Sample FALSE TRUE
donor1_fat 2 156
donor1_skin 19 3784
donor2_fat 24 2794
donor3_fat 297 724
donor3_skin 29 540
donor4_fat 38 50
donor4_skin 47 582
donor5_recovery 122 4757
donor6_fat 25 1969
donor6_skin 14 1696
donor7_fat 26 1681
donor7_skin 51 2847options(repr.plot.width=20, repr.plot.height=20)
ggplot(df, aes(x=col1a1+jam2+prgfrb, y=ccl21+lyve1+prox1+pdpn+cxcl2+ccl2, shape=keep)) +
geom_jitter(width=.01, height = .1) +
geom_vline(xintercept = .05) +
facet_wrap(~Sample) + geom_density_2d()Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”
Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”
Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”
Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”
Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”
Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”
Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”
Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”
Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”
Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”
Warning message:
“`stat_contour()`: Zero contours were generated”
Warning message in min(x):
“nessun argomento non-mancante al minimo; si restituisce Inf”
Warning message in max(x):
“nessun argomento non-mancante al massimo; si restituisce -Inf”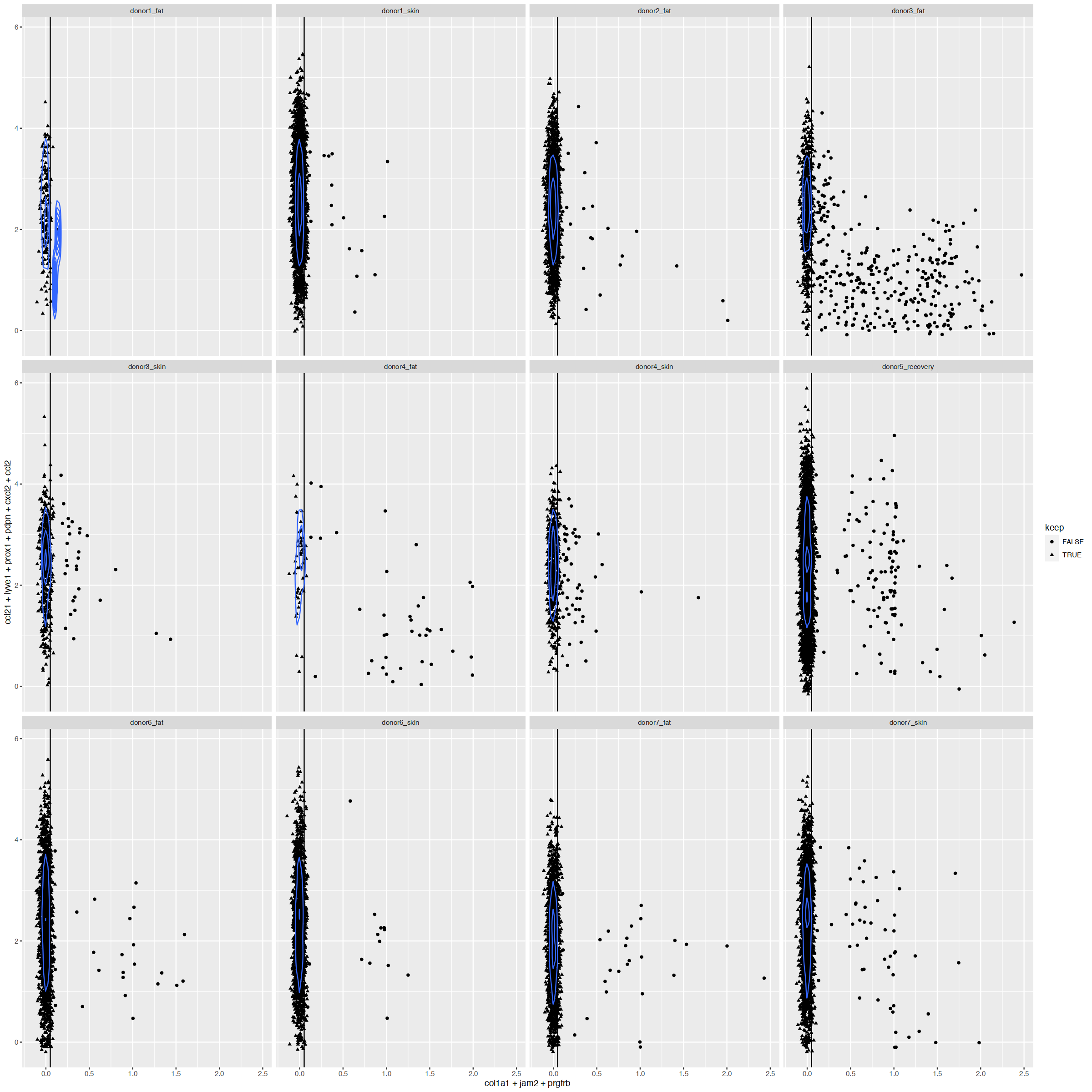
match(rownames(colData(sce)), rownames(df))- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
- 53
- 54
- 55
- 56
- 57
- 58
- 59
- 60
- 61
- 62
- 63
- 64
- 65
- 66
- 67
- 68
- 69
- 70
- 71
- 72
- 73
- 74
- 75
- 76
- 77
- 78
- 79
- 80
- 81
- 82
- 83
- 84
- 85
- 86
- 87
- 88
- 89
- 90
- 91
- 92
- 93
- 94
- 95
- 96
- 97
- 98
- 99
- 100
- 101
- 102
- 103
- 104
- 105
- 106
- 107
- 108
- 109
- 110
- 111
- 112
- 113
- 114
- 115
- 116
- 117
- 118
- 119
- 120
- 121
- 122
- 123
- 124
- 125
- 126
- 127
- 128
- 129
- 130
- 131
- 132
- 133
- 134
- 135
- 136
- 137
- 138
- 139
- 140
- 141
- 142
- 143
- 144
- 145
- 146
- 147
- 148
- 149
- 150
- 151
- 152
- 153
- 154
- 155
- 156
- 157
- 158
- 159
- 160
- 161
- 162
- 163
- 164
- 165
- 166
- 167
- 168
- 169
- 170
- 171
- 172
- 173
- 174
- 175
- 176
- 177
- 178
- 179
- 180
- 181
- 182
- 183
- 184
- 185
- 186
- 187
- 188
- 189
- 190
- 191
- 192
- 193
- 194
- 195
- 196
- 197
- 198
- 199
- 200
- ⋯
- 13766
- 13767
- 13768
- 13769
- 13770
- 13771
- 13772
- 13773
- 13774
- 13775
- 13776
- 13777
- 13778
- 13779
- 13780
- 13781
- 13782
- 13783
- 13784
- 13785
- 13786
- 13787
- 13788
- 13789
- 13790
- 13791
- 13792
- 13793
- 13794
- 13795
- 13796
- 13797
- 13798
- 13799
- 13800
- 13801
- 13802
- 13803
- 13804
- 13805
- 13806
- 13807
- 13808
- 13809
- 13810
- 13811
- 13812
- 13813
- 13814
- 13815
- 13816
- 13817
- 13818
- 13819
- 13820
- 13821
- 13822
- 13823
- 13824
- 13825
- 13826
- 13827
- 13828
- 13829
- 13830
- 13831
- 13832
- 13833
- 13834
- 13835
- 13836
- 13837
- 13838
- 13839
- 13840
- 13841
- 13842
- 13843
- 13844
- 13845
- 13846
- 13847
- 13848
- 13849
- 13850
- 13851
- 13852
- 13853
- 13854
- 13855
- 13856
- 13857
- 13858
- 13859
- 13860
- 13861
- 13862
- 13863
- 13864
- 13865
- 13866
- 13867
- 13868
- 13869
- 13870
- 13871
- 13872
- 13873
- 13874
- 13875
- 13876
- 13877
- 13878
- 13879
- 13880
- 13881
- 13882
- 13883
- 13884
- 13885
- 13886
- 13887
- 13888
- 13889
- 13890
- 13891
- 13892
- 13893
- 13894
- 13895
- 13896
- 13897
- 13898
- 13899
- 13900
- 13901
- 13902
- 13903
- 13904
- 13905
- 13906
- 13907
- 13908
- 13909
- 13910
- 13911
- 13912
- 13913
- 13914
- 13915
- 13916
- 13917
- 13918
- 13919
- 13920
- 13921
- 13922
- 13923
- 13924
- 13925
- 13926
- 13927
- 13928
- 13929
- 13930
- 13931
- 13932
- 13933
- 13934
- 13935
- 13936
- 13937
- 13938
- 13939
- 13940
- 13941
- 13942
- 13943
- 13944
- 13945
- 13946
- 13947
- 13948
- 13949
- 13950
- 13951
- 13952
- 13953
- 13954
- 13955
- 13956
- 13957
- 13958
- 13959
- 13960
- 13961
- 13962
- 13963
- 13964
- 13965
df <- df[match(rownames(colData(sce)), rownames(df)),]
all(rownames(df) == rownames(colData(sce)))
TRUE
sce$outlier_contamination <- !df$keep
sce %>% colData %>% as.data.frame| Sample | Barcode | donor | tissue | tissue_origin | sequencing_run | sex | sum | detected | percent.top_50 | ⋯ | percent.top_200 | percent.top_500 | subsets_Mito_sum | subsets_Mito_detected | subsets_Mito_percent | total | outlier_sum | outlier_detected | sizeFactor | outlier_contamination | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <chr> | <dbl> | <chr> | <chr> | <dbl> | <chr> | <dbl> | <int> | <dbl> | ⋯ | <dbl> | <dbl> | <dbl> | <int> | <dbl> | <dbl> | <otlr.flt> | <otlr.flt> | <dbl> | <lgl> | |
| donor1_fat.AAAGAACCACATACGT-1 | donor1_fat | AAAGAACCACATACGT-1 | 1 | fat | abdomen | 1 | F | 4276 | 1721 | 33.88681 | ⋯ | 52.99345 | 69.31712 | 752 | 16 | 17.5865295 | 4276 | FALSE | FALSE | 0.8543311 | FALSE |
| donor1_fat.AAAGGTAGTCCGGTCA-1 | donor1_fat | AAAGGTAGTCCGGTCA-1 | 1 | fat | abdomen | 1 | F | 3023 | 1084 | 39.66259 | ⋯ | 65.00165 | 80.68144 | 52 | 12 | 1.7201456 | 3023 | FALSE | FALSE | 0.7202661 | FALSE |
| donor1_fat.AAAGGTATCTCTCTTC-1 | donor1_fat | AAAGGTATCTCTCTTC-1 | 1 | fat | abdomen | 1 | F | 3892 | 1422 | 34.12127 | ⋯ | 58.35046 | 75.92497 | 48 | 12 | 1.2332991 | 3892 | FALSE | FALSE | 0.9319094 | FALSE |
| donor1_fat.AACCACACAGTGGCTC-1 | donor1_fat | AACCACACAGTGGCTC-1 | 1 | fat | abdomen | 1 | F | 7722 | 2633 | 26.89718 | ⋯ | 48.67910 | 64.12846 | 103 | 13 | 1.3338513 | 7722 | FALSE | FALSE | 1.8470909 | FALSE |
| donor1_fat.AACGGGATCTGCTGAA-1 | donor1_fat | AACGGGATCTGCTGAA-1 | 1 | fat | abdomen | 1 | F | 3256 | 1284 | 34.33661 | ⋯ | 59.06020 | 75.92138 | 123 | 11 | 3.7776413 | 3256 | FALSE | FALSE | 0.7595401 | FALSE |
| donor1_fat.AAGAACATCTGTGCAA-1 | donor1_fat | AAGAACATCTGTGCAA-1 | 1 | fat | abdomen | 1 | F | 6777 | 1989 | 32.86115 | ⋯ | 60.01180 | 74.07407 | 224 | 14 | 3.3052973 | 6777 | FALSE | FALSE | 1.5886582 | FALSE |
| donor1_fat.AAGCATCTCGATCCCT-1 | donor1_fat | AAGCATCTCGATCCCT-1 | 1 | fat | abdomen | 1 | F | 1746 | 839 | 34.07789 | ⋯ | 61.45475 | 80.58419 | 63 | 11 | 3.6082474 | 1746 | FALSE | FALSE | 0.4080134 | FALSE |
| donor1_fat.AAGCGAGAGTATCCTG-1 | donor1_fat | AAGCGAGAGTATCCTG-1 | 1 | fat | abdomen | 1 | F | 6750 | 2393 | 24.08889 | ⋯ | 46.69630 | 63.71852 | 277 | 18 | 4.1037037 | 6750 | FALSE | FALSE | 1.5692636 | FALSE |
| donor1_fat.AAGTTCGAGGCAGGTT-1 | donor1_fat | AAGTTCGAGGCAGGTT-1 | 1 | fat | abdomen | 1 | F | 5707 | 2041 | 31.20729 | ⋯ | 54.17908 | 68.91537 | 188 | 12 | 3.2942001 | 5707 | FALSE | FALSE | 1.3379833 | FALSE |
| donor1_fat.AATCGACCAATTGCAC-1 | donor1_fat | AATCGACCAATTGCAC-1 | 1 | fat | abdomen | 1 | F | 3401 | 1534 | 26.60982 | ⋯ | 49.51485 | 68.92091 | 107 | 15 | 3.1461335 | 3401 | FALSE | FALSE | 0.7985717 | FALSE |
| donor1_fat.ACAAAGACAATGACCT-1 | donor1_fat | ACAAAGACAATGACCT-1 | 1 | fat | abdomen | 1 | F | 5851 | 1802 | 32.49017 | ⋯ | 58.43446 | 74.67100 | 106 | 16 | 1.8116561 | 5851 | FALSE | FALSE | 1.3927730 | FALSE |
| donor1_fat.ACAAAGACACTAACGT-1 | donor1_fat | ACAAAGACACTAACGT-1 | 1 | fat | abdomen | 1 | F | 3602 | 1004 | 43.80900 | ⋯ | 72.01555 | 86.00777 | 60 | 11 | 1.6657413 | 3602 | FALSE | FALSE | 0.8586948 | FALSE |
| donor1_fat.ACAACCAAGGGCTTCC-1 | donor1_fat | ACAACCAAGGGCTTCC-1 | 1 | fat | abdomen | 1 | F | 10238 | 3273 | 22.24067 | ⋯ | 44.50088 | 59.89451 | 168 | 12 | 1.6409455 | 10238 | FALSE | FALSE | 2.4412922 | FALSE |
| donor1_fat.ACAGGGATCTCATAGG-1 | donor1_fat | ACAGGGATCTCATAGG-1 | 1 | fat | abdomen | 1 | F | 5210 | 1884 | 27.96545 | ⋯ | 53.03263 | 69.17466 | 105 | 13 | 2.0153551 | 5210 | FALSE | FALSE | 1.2376164 | FALSE |
| donor1_fat.ACATTTCCACTCCACT-1 | donor1_fat | ACATTTCCACTCCACT-1 | 1 | fat | abdomen | 1 | F | 3606 | 1305 | 36.30061 | ⋯ | 61.53633 | 77.67610 | 33 | 11 | 0.9151414 | 3606 | FALSE | FALSE | 0.8662102 | FALSE |
| donor1_fat.ACATTTCTCGATGGAG-1 | donor1_fat | ACATTTCTCGATGGAG-1 | 1 | fat | abdomen | 1 | F | 3853 | 1491 | 34.07734 | ⋯ | 58.44796 | 74.27978 | 68 | 13 | 1.7648586 | 3853 | FALSE | FALSE | 0.9176059 | FALSE |
| donor1_fat.ACCCTTGTCACCATAG-1 | donor1_fat | ACCCTTGTCACCATAG-1 | 1 | fat | abdomen | 1 | F | 4627 | 1604 | 34.92544 | ⋯ | 59.39053 | 74.82170 | 84 | 12 | 1.8154312 | 4627 | FALSE | FALSE | 1.1013695 | FALSE |
| donor1_fat.ACCTACCAGACCTCCG-1 | donor1_fat | ACCTACCAGACCTCCG-1 | 1 | fat | abdomen | 1 | F | 1732 | 651 | 46.76674 | ⋯ | 73.78753 | 91.28176 | 41 | 12 | 2.3672055 | 1732 | FALSE | FALSE | 0.4099528 | FALSE |
| donor1_fat.ACGGAAGCACACCTAA-1 | donor1_fat | ACGGAAGCACACCTAA-1 | 1 | fat | abdomen | 1 | F | 5069 | 2279 | 17.65634 | ⋯ | 36.41744 | 56.77648 | 369 | 14 | 7.2795423 | 5069 | FALSE | FALSE | 1.1394313 | FALSE |
| donor1_fat.ACGTAGTTCCCGAAAT-1 | donor1_fat | ACGTAGTTCCCGAAAT-1 | 1 | fat | abdomen | 1 | F | 3897 | 1246 | 36.23300 | ⋯ | 62.63793 | 80.70310 | 39 | 11 | 1.0007698 | 3897 | FALSE | FALSE | 0.9353034 | FALSE |
| donor1_fat.ACGTTCCAGATGAATC-1 | donor1_fat | ACGTTCCAGATGAATC-1 | 1 | fat | abdomen | 1 | F | 1884 | 1136 | 28.60934 | ⋯ | 47.29299 | 66.24204 | 280 | 13 | 14.8619958 | 1884 | FALSE | FALSE | 0.3888612 | FALSE |
| donor1_fat.ACTCTCGAGCCGTTGC-1 | donor1_fat | ACTCTCGAGCCGTTGC-1 | 1 | fat | abdomen | 1 | F | 1220 | 611 | 41.96721 | ⋯ | 66.31148 | 90.90164 | 154 | 15 | 12.6229508 | 1220 | FALSE | FALSE | 0.2584327 | FALSE |
| donor1_fat.ACTTCCGTCAGGAACG-1 | donor1_fat | ACTTCCGTCAGGAACG-1 | 1 | fat | abdomen | 1 | F | 4898 | 2364 | 17.29277 | ⋯ | 34.29971 | 53.14414 | 367 | 16 | 7.4928542 | 4898 | FALSE | FALSE | 1.0984603 | FALSE |
| donor1_fat.AGACACTCAGGCGATA-1 | donor1_fat | AGACACTCAGGCGATA-1 | 1 | fat | abdomen | 1 | F | 4601 | 1518 | 33.51445 | ⋯ | 59.70441 | 76.26603 | 171 | 12 | 3.7165834 | 4601 | FALSE | FALSE | 1.0739746 | FALSE |
| donor1_fat.AGACCATTCAAAGGTA-1 | donor1_fat | AGACCATTCAAAGGTA-1 | 1 | fat | abdomen | 1 | F | 7030 | 2375 | 27.28307 | ⋯ | 50.54054 | 66.28734 | 27 | 10 | 0.3840683 | 7030 | FALSE | FALSE | 1.6977527 | FALSE |
| donor1_fat.AGAGAGCCAAGCCCAC-1 | donor1_fat | AGAGAGCCAAGCCCAC-1 | 1 | fat | abdomen | 1 | F | 3437 | 1047 | 43.26447 | ⋯ | 69.53739 | 84.08496 | 85 | 12 | 2.4730870 | 3437 | FALSE | FALSE | 0.8126327 | FALSE |
| donor1_fat.AGAGCCCAGTCTAGCT-1 | donor1_fat | AGAGCCCAGTCTAGCT-1 | 1 | fat | abdomen | 1 | F | 4781 | 1680 | 33.44489 | ⋯ | 58.12592 | 73.41560 | 110 | 14 | 2.3007739 | 4781 | FALSE | FALSE | 1.1324008 | FALSE |
| donor1_fat.AGATCCAGTGCAGTGA-1 | donor1_fat | AGATCCAGTGCAGTGA-1 | 1 | fat | abdomen | 1 | F | 3119 | 1398 | 30.49054 | ⋯ | 53.83136 | 71.20872 | 84 | 12 | 2.6931709 | 3119 | FALSE | FALSE | 0.7357817 | FALSE |
| donor1_fat.AGATCGTGTTGCGTAT-1 | donor1_fat | AGATCGTGTTGCGTAT-1 | 1 | fat | abdomen | 1 | F | 4444 | 1616 | 32.06571 | ⋯ | 56.79568 | 73.33483 | 7 | 5 | 0.1575158 | 4444 | FALSE | FALSE | 1.0756716 | FALSE |
| donor1_fat.AGGAGGTCAATACGAA-1 | donor1_fat | AGGAGGTCAATACGAA-1 | 1 | fat | abdomen | 1 | F | 3179 | 1211 | 36.83548 | ⋯ | 61.81189 | 77.63448 | 108 | 12 | 3.3972947 | 3179 | FALSE | FALSE | 0.7445093 | FALSE |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋱ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| donor5_recovery.TTTCGATAGTGGAATT-1 | donor5_recovery | TTTCGATAGTGGAATT-1 | 5 | mixed | abdomen | 4 | ? | 3048 | 1575 | 21.42388 | ⋯ | 42.84777 | 64.20604 | 139 | 16 | 4.5603675 | 3048 | FALSE | FALSE | 0.7052353 | FALSE |
| donor5_recovery.TTTCGATGTGCTGATT-1 | donor5_recovery | TTTCGATGTGCTGATT-1 | 5 | mixed | abdomen | 4 | ? | 2238 | 1400 | 17.20286 | ⋯ | 36.90795 | 59.78552 | 57 | 15 | 2.5469169 | 2238 | FALSE | FALSE | 0.5287446 | FALSE |
| donor5_recovery.TTTGACTAGATGGGCT-1 | donor5_recovery | TTTGACTAGATGGGCT-1 | 5 | mixed | abdomen | 4 | ? | 2448 | 1208 | 27.69608 | ⋯ | 53.10458 | 71.07843 | 95 | 16 | 3.8807190 | 2448 | FALSE | FALSE | 0.5704430 | TRUE |
| donor5_recovery.TTTGACTAGTGTTCAC-1 | donor5_recovery | TTTGACTAGTGTTCAC-1 | 5 | mixed | abdomen | 4 | ? | 6499 | 2564 | 24.23450 | ⋯ | 44.72996 | 60.70165 | 78 | 13 | 1.2001846 | 6499 | FALSE | FALSE | 1.5566571 | FALSE |
| donor5_recovery.TTTGACTGTCCAAATC-1 | donor5_recovery | TTTGACTGTCCAAATC-1 | 5 | mixed | abdomen | 4 | ? | 958 | 673 | 24.32150 | ⋯ | 50.62630 | 81.94154 | 8 | 4 | 0.8350731 | 958 | FALSE | FALSE | 0.2303106 | FALSE |
| donor5_recovery.TTTGACTGTGATTCTG-1 | donor5_recovery | TTTGACTGTGATTCTG-1 | 5 | mixed | abdomen | 4 | ? | 1568 | 955 | 26.97704 | ⋯ | 50.70153 | 70.98214 | 71 | 14 | 4.5280612 | 1568 | FALSE | FALSE | 0.3629210 | FALSE |
| donor5_recovery.TTTGACTGTTGTCTAG-1 | donor5_recovery | TTTGACTGTTGTCTAG-1 | 5 | mixed | abdomen | 4 | ? | 936 | 593 | 29.70085 | ⋯ | 58.01282 | 90.06410 | 38 | 11 | 4.0598291 | 936 | FALSE | FALSE | 0.2177041 | FALSE |
| donor5_recovery.TTTGATCAGATGACCG-1 | donor5_recovery | TTTGATCAGATGACCG-1 | 5 | mixed | abdomen | 4 | ? | 956 | 596 | 30.75314 | ⋯ | 58.57741 | 89.95816 | 25 | 8 | 2.6150628 | 956 | FALSE | FALSE | 0.2257044 | FALSE |
| donor5_recovery.TTTGATCAGCAATAAC-1 | donor5_recovery | TTTGATCAGCAATAAC-1 | 5 | mixed | abdomen | 4 | ? | 2148 | 939 | 34.35754 | ⋯ | 61.17318 | 79.56238 | 71 | 14 | 3.3054004 | 2148 | FALSE | FALSE | 0.5035317 | FALSE |
| donor5_recovery.TTTGATCAGTTACTCG-1 | donor5_recovery | TTTGATCAGTTACTCG-1 | 5 | mixed | abdomen | 4 | ? | 3145 | 1486 | 21.90779 | ⋯ | 46.45469 | 67.59936 | 66 | 15 | 2.0985692 | 3145 | FALSE | FALSE | 0.7464487 | FALSE |
| donor5_recovery.TTTGATCGTTGGGCCT-1 | donor5_recovery | TTTGATCGTTGGGCCT-1 | 5 | mixed | abdomen | 4 | ? | 2897 | 1316 | 30.54884 | ⋯ | 54.57370 | 71.83293 | 182 | 17 | 6.2823611 | 2897 | FALSE | FALSE | 0.6582034 | FALSE |
| donor5_recovery.TTTGATCTCAAAGGTA-1 | donor5_recovery | TTTGATCTCAAAGGTA-1 | 5 | mixed | abdomen | 4 | ? | 2155 | 1136 | 26.07889 | ⋯ | 50.06961 | 70.48724 | 98 | 14 | 4.5475638 | 2155 | FALSE | FALSE | 0.4986830 | FALSE |
| donor5_recovery.TTTGGAGAGTGCAAAT-1 | donor5_recovery | TTTGGAGAGTGCAAAT-1 | 5 | mixed | abdomen | 4 | ? | 1054 | 601 | 34.91461 | ⋯ | 61.95446 | 90.41746 | 43 | 14 | 4.0796964 | 1054 | FALSE | FALSE | 0.2450989 | FALSE |
| donor5_recovery.TTTGGAGCATAGGTAA-1 | donor5_recovery | TTTGGAGCATAGGTAA-1 | 5 | mixed | abdomen | 4 | ? | 1846 | 1050 | 23.34778 | ⋯ | 47.50813 | 70.20585 | 89 | 14 | 4.8212351 | 1846 | FALSE | FALSE | 0.4259534 | FALSE |
| donor5_recovery.TTTGGAGGTCAGTTTG-1 | donor5_recovery | TTTGGAGGTCAGTTTG-1 | 5 | mixed | abdomen | 4 | ? | 1222 | 769 | 26.67758 | ⋯ | 53.43699 | 77.98691 | 51 | 13 | 4.1734861 | 1222 | FALSE | FALSE | 0.2838881 | FALSE |
| donor5_recovery.TTTGGAGGTCCAAGAG-1 | donor5_recovery | TTTGGAGGTCCAAGAG-1 | 5 | mixed | abdomen | 4 | ? | 1829 | 1058 | 27.28267 | ⋯ | 49.64461 | 69.49153 | 28 | 12 | 1.5308912 | 1829 | FALSE | FALSE | 0.4366204 | FALSE |
| donor5_recovery.TTTGGAGGTCGACTTA-1 | donor5_recovery | TTTGGAGGTCGACTTA-1 | 5 | mixed | abdomen | 4 | ? | 2389 | 1430 | 17.91545 | ⋯ | 38.21683 | 61.07158 | 154 | 21 | 6.4462118 | 2389 | FALSE | FALSE | 0.5418360 | FALSE |
| donor5_recovery.TTTGGAGGTTGAGAGC-1 | donor5_recovery | TTTGGAGGTTGAGAGC-1 | 5 | mixed | abdomen | 4 | ? | 1618 | 993 | 21.69345 | ⋯ | 46.66255 | 69.53028 | 38 | 11 | 2.3485785 | 1618 | FALSE | FALSE | 0.3830429 | FALSE |
| donor5_recovery.TTTGGAGTCCAAGAGG-1 | donor5_recovery | TTTGGAGTCCAAGAGG-1 | 5 | mixed | abdomen | 4 | ? | 1262 | 830 | 23.85103 | ⋯ | 50.07924 | 73.85103 | 69 | 14 | 5.4675119 | 1262 | FALSE | FALSE | 0.2892216 | FALSE |
| donor5_recovery.TTTGGAGTCTCAACCC-1 | donor5_recovery | TTTGGAGTCTCAACCC-1 | 5 | mixed | abdomen | 4 | ? | 1135 | 717 | 29.51542 | ⋯ | 54.44934 | 80.88106 | 18 | 10 | 1.5859031 | 1135 | FALSE | FALSE | 0.2707968 | FALSE |
| donor5_recovery.TTTGGTTAGCCTCTTC-1 | donor5_recovery | TTTGGTTAGCCTCTTC-1 | 5 | mixed | abdomen | 4 | ? | 763 | 584 | 24.90170 | ⋯ | 49.67235 | 88.99083 | 24 | 8 | 3.1454784 | 763 | FALSE | FALSE | 0.1791574 | FALSE |
| donor5_recovery.TTTGGTTCACTCTAGA-1 | donor5_recovery | TTTGGTTCACTCTAGA-1 | 5 | mixed | abdomen | 4 | ? | 812 | 570 | 27.58621 | ⋯ | 54.43350 | 91.37931 | 24 | 8 | 2.9556650 | 812 | FALSE | FALSE | 0.1910366 | FALSE |
| donor5_recovery.TTTGGTTGTACGTGAG-1 | donor5_recovery | TTTGGTTGTACGTGAG-1 | 5 | mixed | abdomen | 4 | ? | 755 | 494 | 30.99338 | ⋯ | 61.05960 | 100.00000 | 34 | 9 | 4.5033113 | 755 | FALSE | FALSE | 0.1747936 | FALSE |
| donor5_recovery.TTTGGTTGTCCTACAA-1 | donor5_recovery | TTTGGTTGTCCTACAA-1 | 5 | mixed | abdomen | 4 | ? | 835 | 625 | 24.07186 | ⋯ | 49.10180 | 85.02994 | 27 | 6 | 3.2335329 | 835 | FALSE | FALSE | 0.1958852 | FALSE |
| donor5_recovery.TTTGGTTGTGACTCTA-1 | donor5_recovery | TTTGGTTGTGACTCTA-1 | 5 | mixed | abdomen | 4 | ? | 938 | 572 | 29.63753 | ⋯ | 60.34115 | 92.32409 | 2 | 2 | 0.2132196 | 938 | FALSE | FALSE | 0.2269165 | FALSE |
| donor5_recovery.TTTGGTTTCGACCAAT-1 | donor5_recovery | TTTGGTTTCGACCAAT-1 | 5 | mixed | abdomen | 4 | ? | 1328 | 670 | 34.33735 | ⋯ | 64.30723 | 87.19880 | 67 | 10 | 5.0451807 | 1328 | FALSE | FALSE | 0.3057070 | FALSE |
| donor5_recovery.TTTGTTGGTCCTACGG-1 | donor5_recovery | TTTGTTGGTCCTACGG-1 | 5 | mixed | abdomen | 4 | ? | 795 | 552 | 28.30189 | ⋯ | 55.72327 | 93.45912 | 44 | 12 | 5.5345912 | 795 | FALSE | FALSE | 0.1820666 | FALSE |
| donor5_recovery.TTTGTTGGTCGCAGTC-1 | donor5_recovery | TTTGTTGGTCGCAGTC-1 | 5 | mixed | abdomen | 4 | ? | 1872 | 1250 | 16.18590 | ⋯ | 37.01923 | 59.93590 | 66 | 14 | 3.5256410 | 1872 | FALSE | FALSE | 0.4378325 | FALSE |
| donor5_recovery.TTTGTTGGTGGCGTAA-1 | donor5_recovery | TTTGTTGGTGGCGTAA-1 | 5 | mixed | abdomen | 4 | ? | 1928 | 931 | 33.14315 | ⋯ | 58.97303 | 77.64523 | 51 | 13 | 2.6452282 | 1928 | FALSE | FALSE | 0.4550452 | FALSE |
| donor5_recovery.TTTGTTGTCGGCATTA-1 | donor5_recovery | TTTGTTGTCGGCATTA-1 | 5 | mixed | abdomen | 4 | ? | 2091 | 1206 | 21.95122 | ⋯ | 45.28934 | 66.23625 | 53 | 14 | 2.5346724 | 2091 | FALSE | FALSE | 0.4940768 | FALSE |
d <- d[match(rownames(colData(sce)), rownames(d)),]
d$keep <- df$keep
all(rownames(d) == rownames(df))
TRUE
df1 <- cbind(d, df[,-c(1,11)])
df1| rowname | Sample | UMAP.1 | UMAP.2 | ENSG00000117707.PROX1 | ENSG00000162493.PDPN | ENSG00000133800.LYVE1 | ENSG00000137077.CCL21 | ENSG00000189056.RELN | ENSG00000129048.ACKR4 | ⋯ | keep | col1a1 | jam2 | prgfrb | ccl21 | lyve1 | prox1 | pdpn | ccl2 | cxcl2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <chr> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | ⋯ | <lgl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | |
| donor1_fat.AAAGAACCACATACGT-1 | donor1_fat.AAAGAACCACATACGT-1 | donor1_fat | -2.0484646 | -1.1637262 | 1.1180317 | 0.0000000 | 1.7402855 | 6.106210 | 0.0000000 | 0.000000 | ⋯ | TRUE | -6.177371e-03 | -0.0031744491 | 0.025607984 | 0.8637877 | 0.506185147 | 0.399910959 | -2.141122e-02 | -0.0147191045 | -0.0092179474 |
| donor1_fat.AAAGGTAGTCCGGTCA-1 | donor1_fat.AAAGGTAGTCCGGTCA-1 | donor1_fat | 2.8242526 | -1.0391492 | 0.0000000 | 0.0000000 | 0.0000000 | 5.158232 | 0.0000000 | 0.000000 | ⋯ | TRUE | -2.562640e-02 | 0.0135274094 | 0.016411063 | 0.6995753 | -0.030954370 | -0.020318782 | 1.272878e-02 | 0.0321146768 | -0.0155214929 |
| donor1_fat.AAAGGTATCTCTCTTC-1 | donor1_fat.AAAGGTATCTCTCTTC-1 | donor1_fat | 0.9381566 | 0.6268698 | 1.0517659 | 2.0769685 | 0.0000000 | 7.544117 | 0.0000000 | 0.000000 | ⋯ | TRUE | 3.954838e-03 | -0.0047302147 | -0.058921807 | 1.0064774 | 0.009121395 | 0.403507005 | 9.707439e-01 | 0.0009019757 | -0.0097793427 |
| donor1_fat.AACCACACAGTGGCTC-1 | donor1_fat.AACCACACAGTGGCTC-1 | donor1_fat | -0.6080225 | -0.6364202 | 0.0000000 | 0.6242337 | 2.0869030 | 7.002557 | 0.6242337 | 0.000000 | ⋯ | TRUE | -8.390417e-04 | -0.0049775072 | 0.004997854 | 0.9798234 | 0.630962393 | -0.022863151 | 3.719339e-01 | 0.6664521861 | 0.0467031244 |
| donor1_fat.AACGGGATCTGCTGAA-1 | donor1_fat.AACGGGATCTGCTGAA-1 | donor1_fat | -1.0524757 | -1.6120028 | 0.0000000 | 0.0000000 | 3.1537271 | 7.438566 | 1.2120004 | 0.000000 | ⋯ | TRUE | -3.294729e-02 | 0.0172184506 | 0.025801842 | 1.0099529 | 0.952636547 | 0.016039127 | 3.159890e-02 | -0.0161948350 | 0.0093164554 |
| donor1_fat.AAGAACATCTGTGCAA-1 | donor1_fat.AAGAACATCTGTGCAA-1 | donor1_fat | 1.5718066 | 0.7489513 | 2.0521759 | 0.0000000 | 0.0000000 | 4.639207 | 0.0000000 | 0.000000 | ⋯ | TRUE | -9.278279e-03 | -0.0432904238 | -0.004928558 | 0.6361269 | 0.021998822 | 0.764609556 | 7.688593e-03 | 0.4987814014 | 0.2286961444 |
| donor1_fat.AAGCATCTCGATCCCT-1 | donor1_fat.AAGCATCTCGATCCCT-1 | donor1_fat | 1.7909548 | 0.8797356 | 1.7869727 | 0.0000000 | 0.0000000 | 4.182398 | 0.0000000 | 0.000000 | ⋯ | TRUE | 8.961247e-03 | 0.0257253419 | -0.007706475 | 0.5688781 | 0.011359943 | 0.668748819 | -9.337751e-04 | -0.0031576763 | -0.0033508996 |
| donor1_fat.AAGCGAGAGTATCCTG-1 | donor1_fat.AAGCGAGAGTATCCTG-1 | donor1_fat | -1.0558064 | 1.0522912 | 2.0656439 | 0.0000000 | 2.0656439 | 5.340170 | 1.1855387 | 0.000000 | ⋯ | TRUE | -2.025285e-02 | -0.0427843370 | 0.043382435 | 0.7318062 | 0.612130896 | 0.729646983 | 4.289729e-03 | 1.0105701492 | 0.5701520444 |
| donor1_fat.AAGTTCGAGGCAGGTT-1 | donor1_fat.AAGTTCGAGGCAGGTT-1 | donor1_fat | -1.0539844 | -0.7540492 | 0.8052045 | 1.3189166 | 3.0830320 | 5.494934 | 0.0000000 | 0.000000 | ⋯ | TRUE | -1.957310e-02 | -0.0155224185 | -0.014063269 | 0.7817754 | 0.948710864 | 0.299500667 | 7.823209e-01 | -0.0224818197 | -0.0198374980 |
| donor1_fat.AATCGACCAATTGCAC-1 | donor1_fat.AATCGACCAATTGCAC-1 | donor1_fat | -1.3530891 | -1.1469282 | 2.2499633 | 0.0000000 | 1.8091969 | 4.835378 | 0.0000000 | 0.000000 | ⋯ | TRUE | 5.040912e-03 | -0.0047053116 | -0.001473258 | 0.6704721 | 0.547458265 | 0.829585304 | -8.579479e-05 | -0.0042205833 | -0.0122900824 |
| donor1_fat.ACAAAGACAATGACCT-1 | donor1_fat.ACAAAGACAATGACCT-1 | donor1_fat | 1.8577221 | 0.7200912 | 0.7807234 | 0.7807234 | 0.0000000 | 5.148268 | 0.0000000 | 0.000000 | ⋯ | TRUE | -2.382496e-02 | 0.0412669633 | -0.004748681 | 0.7122134 | -0.008654046 | 0.306771378 | 4.814842e-01 | 0.3609760289 | 0.5507497536 |
| donor1_fat.ACAAAGACACTAACGT-1 | donor1_fat.ACAAAGACACTAACGT-1 | donor1_fat | 1.6504609 | -0.9285525 | 0.0000000 | 0.0000000 | 0.0000000 | 1.735139 | 0.0000000 | 0.000000 | ⋯ | TRUE | -2.887057e-02 | -0.0007119733 | -0.011167208 | 0.2209485 | -0.004722751 | -0.006377754 | 1.007664e-02 | -0.0018593270 | 0.9934970903 |
| donor1_fat.ACAACCAAGGGCTTCC-1 | donor1_fat.ACAACCAAGGGCTTCC-1 | donor1_fat | -0.6354316 | 1.3237580 | 1.6079082 | 0.8633345 | 2.7516068 | 5.683224 | 0.8633345 | 0.000000 | ⋯ | TRUE | 7.492564e-03 | 0.0063471223 | 0.009616054 | 0.7539768 | 0.873148301 | 0.580555478 | 5.203047e-01 | 0.3668938136 | -0.0005466140 |
| donor1_fat.ACAGGGATCTCATAGG-1 | donor1_fat.ACAGGGATCTCATAGG-1 | donor1_fat | -0.1048098 | 0.9759096 | 2.5479505 | 0.0000000 | 1.3873679 | 5.628782 | 0.0000000 | 0.000000 | ⋯ | TRUE | 2.690249e-02 | -0.0020851500 | 0.010444795 | 0.7800630 | 0.400894972 | 0.965109975 | -1.875359e-02 | 0.6930383825 | -0.0011001914 |
| donor1_fat.ACATTTCCACTCCACT-1 | donor1_fat.ACATTTCCACTCCACT-1 | donor1_fat | 2.0356176 | -0.3929195 | 0.0000000 | 0.0000000 | 1.1073224 | 7.776050 | 0.0000000 | 1.107322 | ⋯ | TRUE | -2.288523e-02 | 0.0277930016 | -0.001592410 | 1.0105555 | 0.331541713 | -0.017978599 | 2.130233e-02 | 0.9701081869 | 0.6099380962 |
| donor1_fat.ACATTTCTCGATGGAG-1 | donor1_fat.ACATTTCTCGATGGAG-1 | donor1_fat | 0.4631455 | 1.2451497 | 0.0000000 | 0.0000000 | 2.6890671 | 5.406405 | 0.0000000 | 0.000000 | ⋯ | TRUE | 1.469623e-02 | 0.0315918627 | -0.030168437 | 0.7318597 | 0.807769612 | 0.013928482 | 2.950614e-02 | 0.8370519554 | 0.0040403227 |
| donor1_fat.ACCCTTGTCACCATAG-1 | donor1_fat.ACCCTTGTCACCATAG-1 | donor1_fat | 0.8288960 | -0.8372497 | 0.9320313 | 0.0000000 | 1.4936069 | 6.744583 | 1.4936069 | 0.000000 | ⋯ | TRUE | -2.389362e-02 | -0.0308304543 | 0.003702698 | 0.9223370 | 0.451392924 | 0.348397754 | 1.515303e-02 | 1.0203946074 | 0.6950916051 |
| donor1_fat.ACCTACCAGACCTCCG-1 | donor1_fat.ACCTACCAGACCTCCG-1 | donor1_fat | 2.0905075 | 0.3380302 | 0.0000000 | 0.0000000 | 0.0000000 | 8.245428 | 0.0000000 | 0.000000 | ⋯ | TRUE | -5.153461e-05 | -0.0112837207 | -0.016008569 | 1.0031394 | -0.006312337 | -0.006464281 | 3.518851e-03 | 1.0086026451 | -0.0328234172 |
| donor1_fat.ACGGAAGCACACCTAA-1 | donor1_fat.ACGGAAGCACACCTAA-1 | donor1_fat | -3.9451654 | 0.2357324 | 2.1732948 | 0.0000000 | 3.0037904 | 5.174136 | 2.8366140 | 0.000000 | ⋯ | TRUE | -4.068141e-02 | -0.0195332344 | -0.013722141 | 0.7616221 | 0.903429875 | 0.811012906 | -2.210629e-02 | 0.0126439890 | -0.0003801494 |
| donor1_fat.ACGTAGTTCCCGAAAT-1 | donor1_fat.ACGTAGTTCCCGAAAT-1 | donor1_fat | 2.7871909 | 0.2391406 | 0.0000000 | 0.0000000 | 2.3996324 | 6.304685 | 0.0000000 | 0.000000 | ⋯ | TRUE | 1.864060e-02 | 0.0445714489 | 0.006365741 | 0.8880584 | 0.704812734 | -0.033339004 | 1.545340e-03 | 0.5677575058 | -0.0066729671 |
| donor1_fat.ACGTTCCAGATGAATC-1 | donor1_fat.ACGTTCCAGATGAATC-1 | donor1_fat | -3.5143168 | 0.4605635 | 0.0000000 | 0.0000000 | 4.2480247 | 6.801925 | 1.8365751 | 0.000000 | ⋯ | TRUE | -2.603773e-02 | 0.0030491045 | 0.006140261 | 0.9205795 | 0.967926311 | 0.041954248 | 1.651663e-02 | -0.0126926382 | 0.0182637707 |
| donor1_fat.ACTCTCGAGCCGTTGC-1 | donor1_fat.ACTCTCGAGCCGTTGC-1 | donor1_fat | -0.4711578 | -0.9177226 | 0.0000000 | 0.0000000 | 0.0000000 | 6.292590 | 0.0000000 | 0.000000 | ⋯ | TRUE | -1.347052e-02 | -0.0266749866 | -0.005039600 | 0.8886155 | -0.016309032 | 0.003981775 | 4.496706e-02 | 0.0007821924 | -0.0156276358 |
| donor1_fat.ACTTCCGTCAGGAACG-1 | donor1_fat.ACTTCCGTCAGGAACG-1 | donor1_fat | -4.0332029 | 0.4111551 | 2.6920234 | 1.4960688 | 0.0000000 | 3.681983 | 0.9338485 | 0.000000 | ⋯ | TRUE | 3.204020e-02 | -0.0264540226 | 0.001974596 | 0.5470836 | -0.025112726 | 1.016290121 | 8.666760e-01 | 0.0273618310 | -0.0189019862 |
| donor1_fat.AGACACTCAGGCGATA-1 | donor1_fat.AGACACTCAGGCGATA-1 | donor1_fat | 2.8228720 | -1.0549392 | 0.0000000 | 1.5171453 | 1.5171453 | 4.150574 | 0.0000000 | 0.000000 | ⋯ | TRUE | -1.730093e-02 | 0.0059739468 | 0.015334581 | 0.5701491 | 0.476474856 | -0.016580222 | 9.025668e-01 | -0.0097566128 | 0.0045944003 |
| donor1_fat.AGACCATTCAAAGGTA-1 | donor1_fat.AGACCATTCAAAGGTA-1 | donor1_fat | -0.2053631 | 0.8217293 | 1.4683445 | 0.6681318 | 1.1230224 | 5.770836 | 0.0000000 | 0.000000 | ⋯ | TRUE | -1.537451e-02 | -0.0036618865 | 0.010825460 | 0.8017194 | 0.346596916 | 0.572483367 | 4.284791e-01 | 0.5251782663 | 0.6016392301 |
| donor1_fat.AGAGAGCCAAGCCCAC-1 | donor1_fat.AGAGAGCCAAGCCCAC-1 | donor1_fat | 2.1387318 | -0.4795322 | 0.0000000 | 0.0000000 | 1.7912458 | 7.487297 | 0.0000000 | 0.000000 | ⋯ | TRUE | -2.320340e-02 | -0.0068700812 | -0.003882135 | 0.9658855 | 0.574961429 | -0.007573551 | -2.751537e-02 | 1.0131715862 | -0.0037450447 |
| donor1_fat.AGAGCCCAGTCTAGCT-1 | donor1_fat.AGAGCCCAGTCTAGCT-1 | donor1_fat | -0.9772060 | -1.4272784 | 0.9130940 | 0.0000000 | 2.6550031 | 5.973050 | 0.0000000 | 0.000000 | ⋯ | TRUE | 9.452643e-03 | 0.0290292370 | -0.018340308 | 0.8366796 | 0.782972925 | 0.312903236 | 7.415997e-03 | -0.0047334983 | -0.0152212498 |
| donor1_fat.AGATCCAGTGCAGTGA-1 | donor1_fat.AGATCCAGTGCAGTGA-1 | donor1_fat | -1.1057518 | -1.5379166 | 2.6862529 | 0.0000000 | 2.6862529 | 4.745392 | 1.2382358 | 0.000000 | ⋯ | TRUE | -1.201114e-02 | 0.0098563370 | 0.007610442 | 0.6463695 | 0.845765201 | 0.988111397 | -1.628989e-02 | -0.0451875636 | 0.4230879417 |
| donor1_fat.AGATCGTGTTGCGTAT-1 | donor1_fat.AGATCGTGTTGCGTAT-1 | donor1_fat | 0.3803560 | 0.7960730 | 1.5156637 | 1.5156637 | 0.9483405 | 6.802562 | 0.0000000 | 0.000000 | ⋯ | TRUE | 8.615362e-03 | 0.0034245307 | 0.003521553 | 0.9543483 | 0.258806687 | 0.545596044 | 8.743513e-01 | 0.8007611046 | 0.0106937511 |
| donor1_fat.AGGAGGTCAATACGAA-1 | donor1_fat.AGGAGGTCAATACGAA-1 | donor1_fat | -0.5325592 | -1.5191154 | 0.0000000 | 0.0000000 | 2.3304149 | 7.858134 | 0.0000000 | 0.000000 | ⋯ | TRUE | 1.324703e-02 | -0.0081669716 | -0.002687446 | 1.0038940 | 0.705750068 | 0.024315192 | 2.940806e-03 | 0.0233627376 | 0.0038278500 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋱ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| donor5_recovery.TTTCGATAGTGGAATT-1 | donor5_recovery.TTTCGATAGTGGAATT-1 | donor5_recovery | 0.931899659 | 0.9874523770 | 1.273794 | 1.939578 | 2.738090 | 5.295910 | 1.273794 | 0 | ⋯ | TRUE | 0.0078565286 | -0.057756457 | -0.014375509 | 0.72361653 | 8.451639e-01 | 0.465701289 | 1.0021322779 | 0.024510657 | 0.001530271 |
| donor5_recovery.TTTCGATGTGCTGATT-1 | donor5_recovery.TTTCGATGTGCTGATT-1 | donor5_recovery | 3.102502457 | 0.6960900914 | 3.386309 | 0.000000 | 2.257778 | 4.780195 | 1.531704 | 0 | ⋯ | TRUE | -0.0088987663 | 0.005490213 | -0.043369000 | 0.67533106 | 7.024431e-01 | 1.043129369 | -0.0101730319 | -0.014094991 | 0.018495620 |
| donor5_recovery.TTTGACTAGATGGGCT-1 | donor5_recovery.TTTGACTAGATGGGCT-1 | donor5_recovery | -2.869293698 | -1.1133072484 | 0.000000 | 0.000000 | 0.000000 | 5.878965 | 1.461017 | 0 | ⋯ | FALSE | -0.0080437276 | 0.746968267 | -0.018369823 | 0.79137227 | 2.986268e-02 | 0.017854879 | 0.0294468781 | 1.002458934 | 0.955267980 |
| donor5_recovery.TTTGACTAGTGTTCAC-1 | donor5_recovery.TTTGACTAGTGTTCAC-1 | donor5_recovery | 1.664190642 | -1.3018784631 | 2.074509 | 0.000000 | 2.618055 | 5.711280 | 1.192071 | 0 | ⋯ | TRUE | -0.0035028445 | -0.009318380 | 0.003125337 | 0.79445088 | 8.138261e-01 | 0.765391191 | 0.0433107484 | 0.004387637 | 0.021394056 |
| donor5_recovery.TTTGACTGTCCAAATC-1 | donor5_recovery.TTTGACTGTCCAAATC-1 | donor5_recovery | -0.966822632 | -0.6384149898 | 3.275592 | 0.000000 | 0.000000 | 5.730737 | 3.275592 | 0 | ⋯ | TRUE | 0.0354439677 | 0.023959903 | -0.042248876 | 0.79037410 | 1.087581e-03 | 0.993731390 | 0.0034982672 | 0.665586256 | 0.829429226 |
| donor5_recovery.TTTGACTGTGATTCTG-1 | donor5_recovery.TTTGACTGTGATTCTG-1 | donor5_recovery | -0.445122727 | -0.0006952394 | 1.908975 | 1.908975 | 2.702844 | 5.090220 | 0.000000 | 0 | ⋯ | TRUE | -0.0157393377 | -0.008596564 | -0.018597758 | 0.71798386 | 7.873196e-01 | 0.712328288 | 1.0224097005 | 0.530886129 | 0.635044719 |
| donor5_recovery.TTTGACTGTTGTCTAG-1 | donor5_recovery.TTTGACTGTTGTCTAG-1 | donor5_recovery | 1.368694297 | -0.5846982110 | 0.000000 | 2.483723 | 0.000000 | 4.835941 | 2.483723 | 0 | ⋯ | TRUE | -0.0048798873 | 0.041886363 | 0.010982527 | 0.69210490 | 3.078261e-02 | -0.003647282 | 1.0260963791 | 0.663098789 | 1.006265200 |
| donor5_recovery.TTTGATCAGATGACCG-1 | donor5_recovery.TTTGATCAGATGACCG-1 | donor5_recovery | -2.969551571 | -1.9333430637 | 0.000000 | 0.000000 | 0.000000 | 4.785731 | 0.000000 | 0 | ⋯ | TRUE | 0.0049796262 | -0.017490582 | -0.028089584 | 0.71132958 | -8.360597e-05 | -0.011483696 | -0.0162825909 | 0.980354720 | 0.991351882 |
| donor5_recovery.TTTGATCAGCAATAAC-1 | donor5_recovery.TTTGATCAGCAATAAC-1 | donor5_recovery | -1.709560879 | 0.5504284989 | 0.000000 | 0.000000 | 2.313810 | 3.897415 | 2.313810 | 0 | ⋯ | TRUE | 0.0020095366 | -0.016285486 | -0.040712249 | 0.53686022 | 7.452480e-01 | 0.025949401 | 0.0056907374 | 0.406526968 | 0.555830859 |
| donor5_recovery.TTTGATCAGTTACTCG-1 | donor5_recovery.TTTGATCAGTTACTCG-1 | donor5_recovery | 0.267802707 | 1.6214266907 | 1.226309 | 1.226309 | 1.879452 | 4.202872 | 0.000000 | 0 | ⋯ | TRUE | -0.0258017198 | -0.029121559 | -0.003151891 | 0.57797358 | 6.069127e-01 | 0.433515779 | 0.7166358416 | 0.035210088 | 0.996010501 |
| donor5_recovery.TTTGATCGTTGGGCCT-1 | donor5_recovery.TTTGATCGTTGGGCCT-1 | donor5_recovery | -2.176621445 | 0.4464715611 | 2.474530 | 1.333016 | 2.013846 | 5.040236 | 0.000000 | 0 | ⋯ | TRUE | 0.0134119960 | -0.010139829 | -0.005379385 | 0.68251086 | 6.489346e-01 | 0.928533940 | 0.8013680439 | 0.004283741 | -0.002875205 |
| donor5_recovery.TTTGATCTCAAAGGTA-1 | donor5_recovery.TTTGATCTCAAAGGTA-1 | donor5_recovery | -0.603514918 | 1.1894396435 | 2.324973 | 0.000000 | 1.587500 | 5.618437 | 1.587500 | 0 | ⋯ | TRUE | 0.0316904299 | 0.031706872 | -0.005089500 | 0.76535833 | 4.907608e-01 | 0.835413458 | 0.0452100049 | 0.458816864 | 0.529348538 |
| donor5_recovery.TTTGGAGAGTGCAAAT-1 | donor5_recovery.TTTGGAGAGTGCAAAT-1 | donor5_recovery | 1.878585807 | -1.7861404825 | 0.000000 | 0.000000 | 0.000000 | 4.885569 | 0.000000 | 0 | ⋯ | TRUE | -0.0619833289 | -0.013402551 | -0.008624484 | 0.66754052 | -1.080015e-02 | -0.016247571 | 0.0161020449 | 0.022663335 | 0.004043271 |
| donor5_recovery.TTTGGAGCATAGGTAA-1 | donor5_recovery.TTTGGAGCATAGGTAA-1 | donor5_recovery | 1.836256198 | 2.0150197636 | 0.000000 | 2.509784 | 1.743159 | 5.899463 | 0.000000 | 0 | ⋯ | TRUE | -0.0301521554 | -0.031969654 | -0.044194671 | 0.85170500 | 5.239608e-01 | 0.001799755 | 1.0241798955 | 0.463552702 | 0.614902318 |
| donor5_recovery.TTTGGAGGTCAGTTTG-1 | donor5_recovery.TTTGGAGGTCAGTTTG-1 | donor5_recovery | -1.074542531 | -0.6555272449 | 0.000000 | 2.177125 | 0.000000 | 5.750545 | 2.177125 | 0 | ⋯ | TRUE | 0.0012958515 | -0.005301574 | 0.006366746 | 0.76019623 | -1.548313e-02 | -0.005096393 | 0.9813391205 | 0.598935412 | 0.031390193 |
| donor5_recovery.TTTGGAGGTCCAAGAG-1 | donor5_recovery.TTTGGAGGTCCAAGAG-1 | donor5_recovery | -0.190672883 | 0.7834137570 | 2.480430 | 0.000000 | 0.000000 | 5.617554 | 0.000000 | 0 | ⋯ | TRUE | 0.0246976849 | 0.006555066 | 0.022130622 | 0.75928545 | -4.184287e-02 | 0.927049462 | 0.0060278824 | 0.871505983 | 0.000916595 |
| donor5_recovery.TTTGGAGGTCGACTTA-1 | donor5_recovery.TTTGGAGGTCGACTTA-1 | donor5_recovery | 3.671599380 | 1.8101390492 | 1.508721 | 0.000000 | 0.000000 | 5.441227 | 1.508721 | 0 | ⋯ | TRUE | 0.0332257688 | -0.004227929 | 0.034628239 | 0.74670302 | -1.086385e-02 | 0.603558678 | 0.0226744277 | 0.011588924 | 0.025319511 |
| donor5_recovery.TTTGGAGGTTGAGAGC-1 | donor5_recovery.TTTGGAGGTTGAGAGC-1 | donor5_recovery | 0.003620616 | 2.9168378007 | 1.852268 | 1.852268 | 0.000000 | 3.142744 | 1.852268 | 0 | ⋯ | TRUE | -0.0006157999 | -0.035827474 | 0.000994279 | 0.43876522 | -2.388770e-02 | 0.682955565 | 0.9690956326 | 0.008550913 | 0.005462183 |
| donor5_recovery.TTTGGAGTCCAAGAGG-1 | donor5_recovery.TTTGGAGTCCAAGAGG-1 | donor5_recovery | 2.601738057 | -0.4427393783 | 2.156253 | 0.000000 | 0.000000 | 3.890469 | 2.984610 | 0 | ⋯ | TRUE | 0.0081313922 | -0.006917426 | -0.004137815 | 0.53481200 | -1.358548e-02 | 0.793416894 | 0.0234968804 | -0.026179733 | -0.006334056 |
| donor5_recovery.TTTGGAGTCTCAACCC-1 | donor5_recovery.TTTGGAGTCTCAACCC-1 | donor5_recovery | -3.085723408 | -1.6553999815 | 0.000000 | 0.000000 | 0.000000 | 5.379235 | 0.000000 | 0 | ⋯ | TRUE | 0.0229661385 | -0.002785685 | 0.014491942 | 0.69413300 | -6.715803e-03 | 0.013639157 | 0.0319976176 | 0.775310750 | 0.996777514 |
| donor5_recovery.TTTGGTTAGCCTCTTC-1 | donor5_recovery.TTTGGTTAGCCTCTTC-1 | donor5_recovery | 0.379168502 | -0.6033170331 | 0.000000 | 0.000000 | 2.718457 | 6.200887 | 0.000000 | 0 | ⋯ | TRUE | 0.0159770391 | -0.030253281 | 0.018827326 | 0.85607541 | 8.387669e-01 | 0.012585263 | 0.0195137018 | 0.011391660 | 0.014805679 |
| donor5_recovery.TTTGGTTCACTCTAGA-1 | donor5_recovery.TTTGGTTCACTCTAGA-1 | donor5_recovery | -1.271802434 | -0.9042818058 | 0.000000 | 0.000000 | 0.000000 | 5.422125 | 0.000000 | 0 | ⋯ | TRUE | 0.0005551034 | 0.002609067 | 0.017852325 | 0.72905845 | -8.366208e-03 | -0.041417373 | 0.0252338117 | -0.002223779 | 0.039848818 |
| donor5_recovery.TTTGGTTGTACGTGAG-1 | donor5_recovery.TTTGGTTGTACGTGAG-1 | donor5_recovery | -1.942956456 | -1.6052285690 | 0.000000 | 0.000000 | 2.748683 | 5.547458 | 0.000000 | 0 | ⋯ | TRUE | -0.0045435839 | 0.010477268 | 0.026323274 | 0.76677997 | 8.295560e-01 | -0.003306779 | -0.0108690302 | 0.029159402 | 0.003339426 |
| donor5_recovery.TTTGGTTGTCCTACAA-1 | donor5_recovery.TTTGGTTGTCCTACAA-1 | donor5_recovery | 0.055927030 | 0.0280046355 | 3.486722 | 0.000000 | 0.000000 | 4.729289 | 0.000000 | 0 | ⋯ | TRUE | -0.0055102803 | -0.029907189 | 0.002991879 | 0.66621842 | 2.914754e-02 | 0.991302344 | 0.0001031865 | 0.014177467 | -0.012870370 |
| donor5_recovery.TTTGGTTGTGACTCTA-1 | donor5_recovery.TTTGGTTGTGACTCTA-1 | donor5_recovery | -2.104454049 | -0.7065999616 | 0.000000 | 0.000000 | 2.434803 | 5.628656 | 2.434803 | 0 | ⋯ | TRUE | 0.0053178274 | -0.006904177 | -0.021327896 | 0.76737950 | 7.614108e-01 | 0.010816247 | -0.0349556397 | -0.013907964 | 0.992275798 |
| donor5_recovery.TTTGGTTTCGACCAAT-1 | donor5_recovery.TTTGGTTTCGACCAAT-1 | donor5_recovery | -3.072308549 | -1.2226327170 | 0.000000 | 0.000000 | 0.000000 | 6.810155 | 0.000000 | 0 | ⋯ | TRUE | -0.0196506085 | -0.020662980 | 0.020700614 | 0.96829368 | 5.692978e-02 | 0.011930542 | -0.0282590883 | -0.011952372 | 0.023237628 |
| donor5_recovery.TTTGTTGGTCCTACGG-1 | donor5_recovery.TTTGTTGGTCCTACGG-1 | donor5_recovery | -0.856789836 | -1.2551037479 | 0.000000 | 0.000000 | 0.000000 | 5.301861 | 0.000000 | 0 | ⋯ | TRUE | -0.0168946830 | -0.002951992 | -0.006812661 | 0.76314051 | 7.248523e-03 | 0.017844289 | -0.0012970226 | 0.710317045 | 0.022999301 |
| donor5_recovery.TTTGTTGGTCGCAGTC-1 | donor5_recovery.TTTGTTGGTCGCAGTC-1 | donor5_recovery | 3.593091539 | 1.3092576157 | 3.634581 | 1.715445 | 1.715445 | 5.230497 | 1.715445 | 0 | ⋯ | TRUE | -0.0171547874 | 0.004613047 | -0.002779767 | 0.77820613 | 5.612141e-01 | 1.015181387 | 1.0078475380 | -0.011442070 | -0.029356296 |
| donor5_recovery.TTTGTTGGTGGCGTAA-1 | donor5_recovery.TTTGTTGGTGGCGTAA-1 | donor5_recovery | 1.460117213 | -2.1077972461 | 2.431668 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0 | ⋯ | TRUE | 0.0033150125 | -0.018099916 | -0.002460468 | -0.00730366 | 5.005984e-03 | 0.865443464 | -0.0463183614 | -0.008029120 | 0.001738280 |
| donor5_recovery.TTTGTTGTCGGCATTA-1 | donor5_recovery.TTTGTTGTCGGCATTA-1 | donor5_recovery | -0.309542426 | 2.9477435242 | 1.596447 | 0.000000 | 0.000000 | 1.596447 | 0.000000 | 0 | ⋯ | TRUE | -0.0403284420 | -0.019161737 | 0.005990958 | 0.20457458 | 7.002154e-03 | 0.603981121 | 0.0025060108 | -0.019188513 | 0.012098517 |
pdf("figures/contam-removal-spotcheck.pdf", width=10, height=5)
ggplot(df1, aes(x=UMAP.1, y=UMAP.2, colour=ccl21+lyve1+prox1+pdpn+cxcl2+ccl2)) +
geom_point() +
scale_colour_gradient(low = "grey80", high = "blue") +
facet_wrap(~Sample, scales = "free") +
theme(legend.position="bottom")
ggplot(df1, aes(x=UMAP.1, y=UMAP.2, colour=col1a1+jam2+prgfrb)) +
geom_point() +
scale_colour_gradient(low = "grey80", high = "red") +
facet_wrap(~Sample, scales = "free") +
theme(legend.position="bottom")
ggplot(df1, aes(x=UMAP.1, y=UMAP.2)) +
geom_point(colour="grey") +
geom_point(data = df1 %>% dplyr::filter(!keep), colour="black") +
facet_wrap(~Sample, scales = "free") +
theme(legend.position="bottom")
dev.off()
pdf: 2
table(sce$Sample, sce$outlier_contamination)
FALSE TRUE
donor1_fat 156 2
donor1_skin 3784 19
donor2_fat 2794 24
donor3_fat 724 297
donor3_skin 540 29
donor4_fat 50 38
donor4_skin 582 47
donor5_recovery 4757 122
donor6_fat 1969 25
donor6_skin 1696 14
donor7_fat 1681 26
donor7_skin 2847 51sce <- sce[,!sce$outlier_contamination]
sceclass: SingleCellExperiment
dim: 18627 21580
metadata(0):
assays(2): counts logcounts
rownames(18627): ENSG00000187634.SAMD11 ENSG00000188976.NOC2L ...
ENSG00000278817.ENSG00000278817 ENSG00000277196.ENSG00000277196
rowData names(3): ID Symbol Type
colnames(21580): donor1_fat.AAAGAACCACATACGT-1
donor1_fat.AAAGGTAGTCCGGTCA-1 ... donor5_recovery.TTTGTTGGTGGCGTAA-1
donor5_recovery.TTTGTTGTCGGCATTA-1
colData names(21): Sample Barcode ... sizeFactor outlier_contamination
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):table(sce$Sample)
donor1_fat donor1_skin donor2_fat donor3_fat donor3_skin
156 3784 2794 724 540
donor4_fat donor4_skin donor5_recovery donor6_fat donor6_skin
50 582 4757 1969 1696
donor7_fat donor7_skin
1681 2847 saveRDS(sce, file="../output/0.2-sce-after-contamination-removal.rds")